The role of retrotransposable elements in ageing and age-associated diseases
- PMID: 34349292
- PMCID: PMC8600649
- DOI: 10.1038/s41586-021-03542-y
The role of retrotransposable elements in ageing and age-associated diseases
Abstract
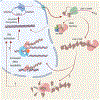
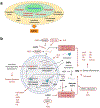
The genomes of virtually all organisms contain repetitive sequences that are generated by the activity of transposable elements (transposons). Transposons are mobile genetic elements that can move from one genomic location to another; in this process, they amplify and increase their presence in genomes, sometimes to very high copy numbers. In this Review we discuss new evidence and ideas that the activity of retrotransposons, a major subgroup of transposons overall, influences and even promotes the process of ageing and age-related diseases in complex metazoan organisms, including humans. Retrotransposons have been coevolving with their host genomes since the dawn of life. This relationship has been largely competitive, and transposons have earned epithets such as 'junk DNA' and 'molecular parasites'. Much of our knowledge of the evolution of retrotransposons reflects their activity in the germline and is evident from genome sequence data. Recent research has provided a wealth of information on the activity of retrotransposons in somatic tissues during an individual lifespan, the molecular mechanisms that underlie this activity, and the manner in which these processes intersect with our own physiology, health and well-being.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Financial Interests.
V.G. and A.S. are cofounders of Persimmon Bio. Inc.; V.G. is SAB member of DoNotAge Inc., Centaura Inc., and Elysium Inc.; J.D.B. is founder of Neochromosome, Inc., founder and director of CDI Labs, Inc., founder and SAB member of ReOpen Diagnostics, and SAB member of Sangamo, Inc., Modern Meadow, Inc., Sample6, Inc. and the Wyss Institute; F.H.G. is SAB member of Transposon Therapeutics, Inc.; J.M.S. is a cofounder and SAB chair of Transposon Therapeutics, Inc. and consults for Atropos Therapeutics, Inc., Gilead Sciences, Inc. and Oncolinea Inc.
Figures

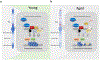
Similar articles
-
The identification of retro-DNAs in primate genomes as DNA transposons mobilizing via retrotransposition.F1000Res. 2024 May 29;12:255. doi: 10.12688/f1000research.130043.3. eCollection 2023. F1000Res. 2024. PMID: 38915770 Free PMC article.
-
Chromosomal distribution and evolution of abundant retrotransposons in plants: gypsy elements in diploid and polyploid Brachiaria forage grasses.Chromosome Res. 2015 Sep;23(3):571-82. doi: 10.1007/s10577-015-9492-6. Chromosome Res. 2015. PMID: 26386563
-
Age-associated de-repression of retrotransposons in the Drosophila fat body, its potential cause and consequence.Aging Cell. 2016 Jun;15(3):542-52. doi: 10.1111/acel.12465. Epub 2016 Apr 12. Aging Cell. 2016. PMID: 27072046 Free PMC article.
-
Becoming a Selfish Clan: Recombination Associated to Reverse-Transcription in LTR Retrotransposons.Genome Biol Evol. 2019 Dec 1;11(12):3382-3392. doi: 10.1093/gbe/evz255. Genome Biol Evol. 2019. PMID: 31755923 Free PMC article. Review.
-
The impact of retrotransposons on human genome evolution.Nat Rev Genet. 2009 Oct;10(10):691-703. doi: 10.1038/nrg2640. Nat Rev Genet. 2009. PMID: 19763152 Free PMC article. Review.
Cited by
-
Artificially stimulating retrotransposon activity increases mortality and accelerates a subset of aging phenotypes in Drosophila.Elife. 2022 Aug 18;11:e80169. doi: 10.7554/eLife.80169. Elife. 2022. PMID: 35980024 Free PMC article.
-
Evidence that conserved essential genes are enriched for pro-longevity factors.Geroscience. 2022 Aug;44(4):1995-2006. doi: 10.1007/s11357-022-00604-5. Epub 2022 Jun 13. Geroscience. 2022. PMID: 35695982 Free PMC article.
-
Kimura's Theory of Non-Adaptive Radiation and Peto's Paradox: A Missing Link?Biology (Basel). 2023 Aug 17;12(8):1140. doi: 10.3390/biology12081140. Biology (Basel). 2023. PMID: 37627024 Free PMC article. Review.
-
Repetitive element transcript accumulation is associated with inflammaging in humans.Geroscience. 2024 Dec;46(6):5663-5679. doi: 10.1007/s11357-024-01126-y. Epub 2024 Apr 20. Geroscience. 2024. PMID: 38641753 Free PMC article.
-
Retrotransposons in embryogenesis and neurodevelopment.Biochem Soc Trans. 2024 Jun 26;52(3):1159-1171. doi: 10.1042/BST20230757. Biochem Soc Trans. 2024. PMID: 38716891 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R37 AG046320/AG/NIA NIH HHS/United States
- R01 AG056306/AG/NIA NIH HHS/United States
- P01 AG047200/AG/NIA NIH HHS/United States
- R01 AG057706/AG/NIA NIH HHS/United States
- K99 AG057812/AG/NIA NIH HHS/United States
- R01 AG024353/AG/NIA NIH HHS/United States
- R01 AG016694/AG/NIA NIH HHS/United States
- RF1 AG024353/AG/NIA NIH HHS/United States
- T32 AG041688/AG/NIA NIH HHS/United States
- R01 AG027237/AG/NIA NIH HHS/United States
- P01 AG051449/AG/NIA NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- R21 CA235521/CA/NCI NIH HHS/United States
- P20 GM119943/GM/NIGMS NIH HHS/United States
- R01 AG067306/AG/NIA NIH HHS/United States
- R37 AG016694/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

