Composition of the Fecal Microbiota of Piglets at Various Growth Stages
- PMID: 34336969
- PMCID: PMC8319241
- DOI: 10.3389/fvets.2021.661671
Composition of the Fecal Microbiota of Piglets at Various Growth Stages
Abstract
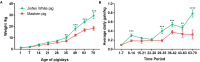
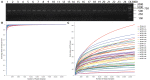
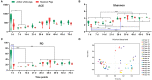
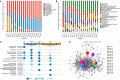
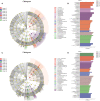
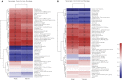
Gastrointestinal (GI) microbiota play an important role in promoting growth in piglets. However, studies on microbiota composition at various growth stages are lacking. We measured body weights of Jinfen White and Mashen piglets every 7 days and collected their fecal samples by rectal swabbing at nine time points during suckling (1-28 days) and nursery (35-70 days) stages to gain insight into microbiota variability during piglet growth. The fecal microbiota were characterized via 16S rRNA gene sequencing to analyze the effects of microbial diversity on piglet growth and development preliminarily. The results showed that although the two breeds of piglets have similar body weights at birth, weaned Jinfen White piglets demonstrated a significantly greater body weight and daily weight gain than weaned Mashen piglets (P < 0.01). A total of 1,976 operational taxonomic units (OTUs) belonging to 27 phyla and 489 genera were uncovered, in which the highest numbers of OTUs belong to the phyla Firmicutes and Bacteroidetes. Lactobacillus, Bacteroides, and Prevotellaceae NK3B31 groups accounting for 12.4, 8.8, and 5.8% of OTUs, respectively, showed relatively high abundance at the genus level. Nine sampling time points were divided into three growth stages, namely, immediate postfarrowing (1 day old), suckling (7, 14, and 21 days old), and nursery (28, 35, 49, 63, and 70 days old), on the basis of the results of microbial diversity, principal coordinate, and co-occurrence network analyses. In addition, it identified 54 discriminative features in the microbiota between two breeds of piglets by LEfSe analysis, in which 17 genera enriched the microbiota community of Jinfen White piglets. Finally, abundances of 29 genera showed significant positive correlations with body weights and daily weight gain of piglets. Conversely, abundances of 12 genera demonstrated significant negative correlations with body weights of piglets. The results of our study will provide a theoretical basis for succession patterns in fecal microbiota of piglets and suggest the need for meticulous management of piglets in pig production.
Keywords: 16S rRNA sequencing; fecal microbiota; growth and development; jinfen white pig; mashen pig; piglet.
Copyright © 2021 Yang, Liu, Liu, Wang, Guo, Du, Cai, Zhao, Lu, Guo, Cao, Duan, Li and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Effects of Maternal Factors and Postpartum Environment on Early Colonization of Intestinal Microbiota in Piglets.Front Vet Sci. 2022 Apr 7;9:815944. doi: 10.3389/fvets.2022.815944. eCollection 2022. Front Vet Sci. 2022. PMID: 35464386 Free PMC article.
-
Comparative Evaluation of the Ileum Microbiota Composition in Piglets at Different Growth Stages.Front Microbiol. 2021 Dec 1;12:765691. doi: 10.3389/fmicb.2021.765691. eCollection 2021. Front Microbiol. 2021. PMID: 34925272 Free PMC article.
-
Impact of Intestinal Microbiota on Growth Performance of Suckling and Weaned Piglets.Microbiol Spectr. 2023 Jun 15;11(3):e0374422. doi: 10.1128/spectrum.03744-22. Epub 2023 Apr 6. Microbiol Spectr. 2023. PMID: 37022154 Free PMC article.
-
Late weaning is associated with increased microbial diversity and Faecalibacterium prausnitzii abundance in the fecal microbiota of piglets.Anim Microbiome. 2020 Jan 16;2(1):2. doi: 10.1186/s42523-020-0020-4. Anim Microbiome. 2020. PMID: 33499995 Free PMC article.
-
The Maturing Development of Gut Microbiota in Commercial Piglets during the Weaning Transition.Front Microbiol. 2017 Sep 4;8:1688. doi: 10.3389/fmicb.2017.01688. eCollection 2017. Front Microbiol. 2017. PMID: 28928724 Free PMC article.
Cited by
-
Effects of Low-Ambient-Temperature Stimulation on Modifying the Intestinal Structure and Function of Different Pig Breeds.Animals (Basel). 2022 Oct 12;12(20):2740. doi: 10.3390/ani12202740. Animals (Basel). 2022. PMID: 36290125 Free PMC article.
-
Effects of Maternal Factors and Postpartum Environment on Early Colonization of Intestinal Microbiota in Piglets.Front Vet Sci. 2022 Apr 7;9:815944. doi: 10.3389/fvets.2022.815944. eCollection 2022. Front Vet Sci. 2022. PMID: 35464386 Free PMC article.
-
Uncovering the mechanism of Clostridium butyricum CBX 2021 to improve pig health based on in vivo and in vitro studies.Front Microbiol. 2024 Jun 14;15:1394332. doi: 10.3389/fmicb.2024.1394332. eCollection 2024. Front Microbiol. 2024. PMID: 38946904 Free PMC article.
-
Comparative Evaluation of the Ileum Microbiota Composition in Piglets at Different Growth Stages.Front Microbiol. 2021 Dec 1;12:765691. doi: 10.3389/fmicb.2021.765691. eCollection 2021. Front Microbiol. 2021. PMID: 34925272 Free PMC article.
-
Clostridium butyricum Prevents Diarrhea Incidence in Weaned Piglets Induced by Escherichia coli K88 through Rectal Bacteria-Host Metabolic Cross-Talk.Animals (Basel). 2024 Aug 6;14(16):2287. doi: 10.3390/ani14162287. Animals (Basel). 2024. PMID: 39199821 Free PMC article.
References
LinkOut - more resources
Full Text Sources

