PRMT1-dependent regulation of RNA metabolism and DNA damage response sustains pancreatic ductal adenocarcinoma
- PMID: 34330913
- PMCID: PMC8324870
- DOI: 10.1038/s41467-021-24798-y
PRMT1-dependent regulation of RNA metabolism and DNA damage response sustains pancreatic ductal adenocarcinoma
Abstract
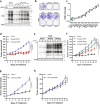
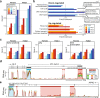
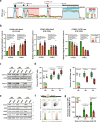
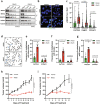
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer that has remained clinically challenging to manage. Here we employ an RNAi-based in vivo functional genomics platform to determine epigenetic vulnerabilities across a panel of patient-derived PDAC models. Through this, we identify protein arginine methyltransferase 1 (PRMT1) as a critical dependency required for PDAC maintenance. Genetic and pharmacological studies validate the role of PRMT1 in maintaining PDAC growth. Mechanistically, using proteomic and transcriptomic analyses, we demonstrate that global inhibition of asymmetric arginine methylation impairs RNA metabolism, which includes RNA splicing, alternative polyadenylation, and transcription termination. This triggers a robust downregulation of multiple pathways involved in the DNA damage response, thereby promoting genomic instability and inhibiting tumor growth. Taken together, our data support PRMT1 as a compelling target in PDAC and informs a mechanism-based translational strategy for future therapeutic development.Statement of significancePDAC is a highly lethal cancer with limited therapeutic options. This study identified and characterized PRMT1-dependent regulation of RNA metabolism and coordination of key cellular processes required for PDAC tumor growth, defining a mechanism-based translational hypothesis for PRMT1 inhibitors.
© 2021. The Author(s).
Conflict of interest statement
G. F. D. reports personal fees from and stock ownership in Karyopharm Therapeutics, Forma Therapeutics, Metabomed, BiovelocITA, Nurix and Orionis Biosciences; and personal fees from Blueprint Medicines, Taiho Pharmaceutical, Symphogen and Helsinn Ventures. T. P. H. reports personal fees and stock ownership from Cullgen Inc. The remaining authors declare no competing interests.
Figures


Similar articles
-
PRMT1 promotes pancreatic cancer growth and predicts poor prognosis.Cell Oncol (Dordr). 2020 Feb;43(1):51-62. doi: 10.1007/s13402-019-00435-1. Epub 2019 Sep 13. Cell Oncol (Dordr). 2020. PMID: 31520395
-
PRMT1 promotes pancreatic cancer development and resistance to chemotherapy.Cell Rep Med. 2024 Mar 19;5(3):101461. doi: 10.1016/j.xcrm.2024.101461. Epub 2024 Mar 8. Cell Rep Med. 2024. PMID: 38460517 Free PMC article.
-
PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma.Theranostics. 2021 Mar 12;11(11):5387-5403. doi: 10.7150/thno.42345. eCollection 2021. Theranostics. 2021. PMID: 33859753 Free PMC article.
-
Protein Arginine Methyltransferases in Pancreatic Ductal Adenocarcinoma: New Molecular Targets for Therapy.Int J Mol Sci. 2024 Apr 2;25(7):3958. doi: 10.3390/ijms25073958. Int J Mol Sci. 2024. PMID: 38612768 Free PMC article. Review.
-
PRMT1 in human neoplasm: cancer biology and potential therapeutic target.Cell Commun Signal. 2024 Feb 8;22(1):102. doi: 10.1186/s12964-024-01506-z. Cell Commun Signal. 2024. PMID: 38326807 Free PMC article. Review.
Cited by
-
PRMT1 driven PTX3 regulates ferritinophagy in glioma.Autophagy. 2023 Jul;19(7):1997-2014. doi: 10.1080/15548627.2023.2165757. Epub 2023 Jan 16. Autophagy. 2023. PMID: 36647288 Free PMC article.
-
Genomic instability genes in lung and colon adenocarcinoma indicate organ specificity of transcriptomic impact on Copy Number Alterations.Sci Rep. 2022 Jul 11;12(1):11739. doi: 10.1038/s41598-022-15692-8. Sci Rep. 2022. PMID: 35817785 Free PMC article.
-
Somatic Mutations in Core Spliceosome Components Promote Tumorigenesis and Generate an Exploitable Vulnerability in Human Cancer.Cancers (Basel). 2022 Apr 4;14(7):1827. doi: 10.3390/cancers14071827. Cancers (Basel). 2022. PMID: 35406598 Free PMC article. Review.
-
PRMT1 and PRMT5: on the road of homologous recombination and non-homologous end joining.Genome Instab Dis. 2023 Aug;4(4):197-209. doi: 10.1007/s42764-022-00095-w. Epub 2022 Dec 7. Genome Instab Dis. 2023. PMID: 37663901 Free PMC article.
-
FACS-based genome-wide CRISPR screens define key regulators of DNA damage signaling pathways.Mol Cell. 2023 Aug 3;83(15):2810-2828.e6. doi: 10.1016/j.molcel.2023.07.004. Mol Cell. 2023. PMID: 37541219 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

