Jasmonate inhibits adventitious root initiation through repression of CKX1 and activation of RAP2.6L transcription factor in Arabidopsis
- PMID: 34329421
- PMCID: PMC8547155
- DOI: 10.1093/jxb/erab358
Jasmonate inhibits adventitious root initiation through repression of CKX1 and activation of RAP2.6L transcription factor in Arabidopsis
Abstract
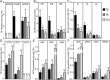
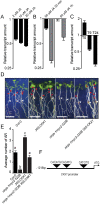
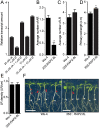
Adventitious rooting is a de novo organogenesis process that enables plants to propagate clonally and cope with environmental stresses. Adventitious root initiation (ARI) is controlled by interconnected transcriptional and hormonal networks, but there is little knowledge of the genetic and molecular programs orchestrating these networks. Thus, we have applied genome-wide transcriptome profiling to elucidate the transcriptional reprogramming events preceding ARI. These reprogramming events are associated with the down-regulation of cytokinin (CK) signaling and response genes, which could be triggers for ARI. Interestingly, we found that CK free base (iP, tZ, cZ, and DHZ) content declined during ARI, due to down-regulation of de novo CK biosynthesis and up-regulation of CK inactivation pathways. We also found that MYC2-dependent jasmonate (JA) signaling inhibits ARI by down-regulating the expression of the CYTOKININ OXIDASE/DEHYDROGENASE1 (CKX1) gene. We also demonstrated that JA and CK synergistically activate expression of the transcription factor RELATED to APETALA2.6 LIKE (RAP2.6L), and constitutive expression of this transcription factor strongly inhibits ARI. Collectively, our findings reveal that previously unknown genetic interactions between JA and CK play key roles in ARI.
Keywords: Adventitious roots; Arabidopsis; CKX1; MYC2; RAP2.6L; cytokinins; jasmonate; light; vegetative propagation.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

Comment in
-
Dual roles of jasmonate in adventitious rooting.J Exp Bot. 2021 Oct 26;72(20):6808-6810. doi: 10.1093/jxb/erab378. J Exp Bot. 2021. PMID: 34698862 Free PMC article.
Similar articles
-
ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis.New Phytol. 2020 Dec;228(5):1611-1626. doi: 10.1111/nph.16794. Epub 2020 Aug 6. New Phytol. 2020. PMID: 32634250
-
A DAO1-Mediated Circuit Controls Auxin and Jasmonate Crosstalk Robustness during Adventitious Root Initiation in Arabidopsis.Int J Mol Sci. 2019 Sep 9;20(18):4428. doi: 10.3390/ijms20184428. Int J Mol Sci. 2019. PMID: 31505771 Free PMC article.
-
Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis.Plant Cell. 2012 Jun;24(6):2515-27. doi: 10.1105/tpc.112.099119. Epub 2012 Jun 22. Plant Cell. 2012. PMID: 22730403 Free PMC article.
-
The plant Mediator complex and its role in jasmonate signaling.J Exp Bot. 2019 Jul 5;70(13):3415-3424. doi: 10.1093/jxb/erz233. J Exp Bot. 2019. PMID: 31089685 Free PMC article. Review.
-
Mediator subunit MED25: at the nexus of jasmonate signaling.Curr Opin Plant Biol. 2020 Oct;57:78-86. doi: 10.1016/j.pbi.2020.06.006. Epub 2020 Aug 7. Curr Opin Plant Biol. 2020. PMID: 32777679 Review.
Cited by
-
Dual roles of jasmonate in adventitious rooting.J Exp Bot. 2021 Oct 26;72(20):6808-6810. doi: 10.1093/jxb/erab378. J Exp Bot. 2021. PMID: 34698862 Free PMC article.
-
The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling.Front Plant Sci. 2022 Apr 25;13:868874. doi: 10.3389/fpls.2022.868874. eCollection 2022. Front Plant Sci. 2022. PMID: 35548315 Free PMC article. Review.
-
Factors governing cellular reprogramming competence in Arabidopsis adventitious root formation.Dev Cell. 2024 Oct 21;59(20):2745-2758.e3. doi: 10.1016/j.devcel.2024.06.019. Epub 2024 Jul 22. Dev Cell. 2024. PMID: 39043189
-
Plant regeneration in the new era: from molecular mechanisms to biotechnology applications.Sci China Life Sci. 2024 Jul;67(7):1338-1367. doi: 10.1007/s11427-024-2581-2. Epub 2024 May 31. Sci China Life Sci. 2024. PMID: 38833085 Review.
-
RNA-Seq Reveals Waterlogging-Triggered Root Plasticity in Mungbean Associated with Ethylene and Jasmonic Acid Signal Integrators for Root Regeneration.Plants (Basel). 2022 Mar 30;11(7):930. doi: 10.3390/plants11070930. Plants (Basel). 2022. PMID: 35406910 Free PMC article.
References
-
- Avalbaev A, Yuldashev R, Fedorova K, Somov K, Vysotskaya L, Allagulova C, Shakirova F. 2016. Exogenous methyl jasmonate regulates cytokinin content by modulating cytokinin oxidase activity in wheat seedlings under salinity. Journal of Plant Physiology 191, 101–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

