Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers
- PMID: 34314702
- PMCID: PMC8380725
- DOI: 10.1016/j.cell.2021.07.005
Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers
Abstract
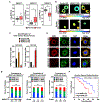
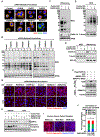
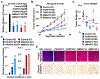
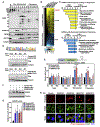
Defects in translation lead to changes in the expression of proteins that can serve as drivers of cancer formation. Here, we show that cytosolic NAD+ synthesis plays an essential role in ovarian cancer by regulating translation and maintaining protein homeostasis. Expression of NMNAT-2, a cytosolic NAD+ synthase, is highly upregulated in ovarian cancers. NMNAT-2 supports the catalytic activity of the mono(ADP-ribosyl) transferase (MART) PARP-16, which mono(ADP-ribosyl)ates (MARylates) ribosomal proteins. Depletion of NMNAT-2 or PARP-16 leads to inhibition of MARylation, increased polysome association and enhanced translation of specific mRNAs, aggregation of their translated protein products, and reduced growth of ovarian cancer cells. Furthermore, MARylation of the ribosomal proteins, such as RPL24 and RPS6, inhibits polysome assembly by stabilizing eIF6 binding to ribosomes. Collectively, our results demonstrate that ribosome MARylation promotes protein homeostasis in cancers by fine-tuning the levels of protein synthesis and preventing toxic protein aggregation.
Keywords: ADP-ribosylation; MARylation; NAD(+); NAD(+) sensor; NAD(+) synthesis; NMNAT-2; PARP-16; cancer; mRNA translation; mono(ADP-ribose); mono(ADP-ribosyl)ation; ovarian cancer; protein aggregation; protein synthesis; ribosomes; translation.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.L.K. is a founder and consultant for Ribon Therapeutics, Inc. and ARase Therapeutics, Inc. He is also coholder of U.S. Patent 9,599,606 covering the ADP-ribose detection reagent used herein, which has been licensed to and is sold by EMD Millipore.
Figures


Similar articles
-
Ribosome ADP-ribosylation: A mechanism for maintaining protein homeostasis in cancers.Cell Biol Int. 2022 Mar;46(3):333-335. doi: 10.1002/cbin.11745. Epub 2021 Dec 17. Cell Biol Int. 2022. PMID: 34897867
-
RACK1 MARylation regulates translation and stress granules in ovarian cancer cells.J Cell Biol. 2025 Feb 3;224(2):e202401101. doi: 10.1083/jcb.202401101. Epub 2025 Jan 6. J Cell Biol. 2025. PMID: 39760726 Free PMC article.
-
Noncanonical mono(ADP-ribosyl)ation of zinc finger SZF proteins counteracts ubiquitination for protein homeostasis in plant immunity.Mol Cell. 2021 Nov 18;81(22):4591-4604.e8. doi: 10.1016/j.molcel.2021.09.006. Epub 2021 Sep 29. Mol Cell. 2021. PMID: 34592134 Free PMC article.
-
MARTs and MARylation in the Cytosol: Biological Functions, Mechanisms of Action, and Therapeutic Potential.Cells. 2021 Feb 3;10(2):313. doi: 10.3390/cells10020313. Cells. 2021. PMID: 33546365 Free PMC article. Review.
-
PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis.Genes Dev. 2020 Mar 1;34(5-6):302-320. doi: 10.1101/gad.334433.119. Epub 2020 Feb 6. Genes Dev. 2020. PMID: 32029452 Free PMC article. Review.
Cited by
-
Exploring the molecular mechanism of hepatitis virus inducing hepatocellular carcinoma by microarray data and immune infiltrates analysis.Front Immunol. 2022 Nov 11;13:1032819. doi: 10.3389/fimmu.2022.1032819. eCollection 2022. Front Immunol. 2022. PMID: 36439183 Free PMC article.
-
Ribosome biogenesis in disease: new players and therapeutic targets.Signal Transduct Target Ther. 2023 Jan 9;8(1):15. doi: 10.1038/s41392-022-01285-4. Signal Transduct Target Ther. 2023. PMID: 36617563 Free PMC article. Review.
-
urPTMdb/TeaProt: Upstream and Downstream Proteomics Analysis.J Proteome Res. 2023 Feb 3;22(2):302-310. doi: 10.1021/acs.jproteome.2c00048. Epub 2022 Jun 27. J Proteome Res. 2023. PMID: 35759515 Free PMC article.
-
Cryptic phosphoribosylase activity of NAMPT restricts the virion incorporation of viral proteins.Nat Metab. 2024 Dec;6(12):2300-2318. doi: 10.1038/s42255-024-01162-0. Epub 2024 Nov 21. Nat Metab. 2024. PMID: 39572750
-
Biological Functions and Therapeutic Potential of NAD+ Metabolism in Gynecological Cancers.Cancers (Basel). 2024 Sep 5;16(17):3085. doi: 10.3390/cancers16173085. Cancers (Basel). 2024. PMID: 39272943 Free PMC article. Review.
References
-
- Andrews S (2010). FASTQC. A quality control tool for high throughput sequence data.
-
- Berger F, Lau C, Dahlmann M, and Ziegler M (2005). Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 280, 36334–36341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

