Asymmetric nuclear division in neural stem cells generates sibling nuclei that differ in size, envelope composition, and chromatin organization
- PMID: 34297912
- PMCID: PMC8491657
- DOI: 10.1016/j.cub.2021.06.063
Asymmetric nuclear division in neural stem cells generates sibling nuclei that differ in size, envelope composition, and chromatin organization
Abstract
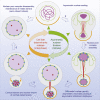
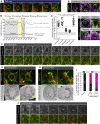
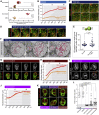
Although nuclei are the defining features of eukaryotes, we still do not fully understand how the nuclear compartment is duplicated and partitioned during division. This is especially the case for organisms that do not completely disassemble their nuclear envelope upon entry into mitosis. In studying this process in Drosophila neural stem cells, which undergo asymmetric divisions, we find that the nuclear compartment boundary persists during mitosis thanks to the maintenance of a supporting nuclear lamina. This mitotic nuclear envelope is then asymmetrically remodeled and partitioned to give rise to two daughter nuclei that differ in envelope composition and exhibit a >30-fold difference in volume. The striking difference in nuclear size was found to depend on two consecutive processes: asymmetric nuclear envelope resealing at mitotic exit at sites defined by the central spindle, and differential nuclear growth that appears to depend on the available local reservoir of ER/nuclear membranes, which is asymmetrically partitioned between the two daughter cells. Importantly, these asymmetries in size and composition of the daughter nuclei, and the associated asymmetries in chromatin organization, all become apparent long before the cortical release and the nuclear import of cell fates determinants. Thus, asymmetric nuclear remodeling during stem cell divisions may contribute to the generation of cellular diversity by initiating distinct transcriptional programs in sibling nuclei that contribute to later changes in daughter cell identity and fate.
Keywords: asymmetric division; cell fate; chromatin; closed mitosis; cytokinesis; nuclear division; nuclear envelope remodelling; open mitosis; spindle; stem cells.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declarations of interests The authors declare no competing interests.
Figures





Comment in
-
Cell biology: How does the nucleus get its membrane?Curr Biol. 2021 Sep 27;31(18):R1077-R1079. doi: 10.1016/j.cub.2021.08.014. Curr Biol. 2021. PMID: 34582813 Free PMC article.
Similar articles
-
The mode of mitosis is dramatically modified by deletion of a single nuclear pore complex gene in Aspergillus nidulans.Fungal Genet Biol. 2019 Sep;130:72-81. doi: 10.1016/j.fgb.2019.04.010. Epub 2019 Apr 23. Fungal Genet Biol. 2019. PMID: 31026588
-
Nuclear envelope remodelling during mitosis.Curr Opin Cell Biol. 2021 Jun;70:67-74. doi: 10.1016/j.ceb.2020.12.004. Epub 2021 Jan 6. Curr Opin Cell Biol. 2021. PMID: 33421755 Free PMC article. Review.
-
Divergence of mitotic strategies in fission yeasts.Nucleus. 2012 May-Jun;3(3):220-5. doi: 10.4161/nucl.19514. Epub 2012 May 1. Nucleus. 2012. PMID: 22572960 Free PMC article.
-
Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo.Eur J Cell Biol. 1984 May;34(1):179-89. Eur J Cell Biol. 1984. PMID: 6428889
-
The nuclear envelope.Cold Spring Harb Perspect Biol. 2010 Mar;2(3):a000539. doi: 10.1101/cshperspect.a000539. Cold Spring Harb Perspect Biol. 2010. PMID: 20300205 Free PMC article. Review.
Cited by
-
Aurora B-dependent polarization of the cortical actomyosin network during mitotic exit.EMBO Rep. 2021 Oct 5;22(10):e52387. doi: 10.15252/embr.202152387. Epub 2021 Aug 24. EMBO Rep. 2021. PMID: 34431205 Free PMC article.
-
Asymmetric chromatin retention and nuclear envelopes separate chromosomes in fused cells in vivo.Commun Biol. 2022 Sep 19;5(1):953. doi: 10.1038/s42003-022-03874-z. Commun Biol. 2022. PMID: 36123528 Free PMC article.
-
The cytokinetic midbody mediates asymmetric fate specification at mitotic exit during neural stem cell division.bioRxiv [Preprint]. 2024 Aug 28:2024.08.27.609974. doi: 10.1101/2024.08.27.609974. bioRxiv. 2024. PMID: 39253494 Free PMC article. Preprint.
-
Cell biology: How does the nucleus get its membrane?Curr Biol. 2021 Sep 27;31(18):R1077-R1079. doi: 10.1016/j.cub.2021.08.014. Curr Biol. 2021. PMID: 34582813 Free PMC article.
-
The Drosophila neuroblast polarity cycle at a glance.J Cell Sci. 2024 Mar 1;137(5):jcs261789. doi: 10.1242/jcs.261789. Epub 2024 Mar 11. J Cell Sci. 2024. PMID: 38465513
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

