CRISPR/Cas9-mediated genome editing reveals 12 testis-enriched genes dispensable for male fertility in mice
- PMID: 34290169
- PMCID: PMC9226692
- DOI: 10.4103/aja.aja_63_21
CRISPR/Cas9-mediated genome editing reveals 12 testis-enriched genes dispensable for male fertility in mice
Abstract
Gene expression analyses suggest that more than 1000-2000 genes are expressed predominantly in mouse and human testes. Although functional analyses of hundreds of these genes have been performed, there are still many testis-enriched genes whose functions remain unexplored. Analyzing gene function using knockout (KO) mice is a powerful tool to discern if the gene of interest is essential for sperm formation, function, and male fertility in vivo. In this study, we generated KO mice for 12 testis-enriched genes, 1700057G04Rik, 4921539E11Rik, 4930558C23Rik, Cby2, Ldhal6b, Rasef, Slc25a2, Slc25a41, Smim8, Smim9, Tmem210, and Tomm20l, using the clustered regularly interspaced short palindromic repeats /CRISPR-associated protein 9 (CRISPR/Cas9) system. We designed two gRNAs for each gene to excise almost all the protein-coding regions to ensure that the deletions in these genes result in a null mutation. Mating tests of KO mice reveal that these 12 genes are not essential for male fertility, at least when individually ablated, and not together with other potentially compensatory paralogous genes. Our results could prevent other laboratories from expending duplicative effort generating KO mice, for which no apparent phenotype exists.
Keywords: CRISPR/Cas9; knockout mice; male infertility; spermatozoa; testis.
Conflict of interest statement
None
Figures

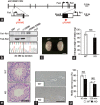
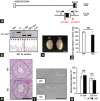
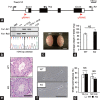
Comment in
-
Commentary on "CRISPR/Cas9-mediated genome editing reveals 12 testis-enriched genes dispensable for male fertility in mice".Asian J Androl. 2022 May-Jun;24(3):273. doi: 10.4103/aja202196. Asian J Androl. 2022. PMID: 34810375 Free PMC article. No abstract available.
Similar articles
-
Individual disruption of 12 testis-enriched genes via the CRISPR/Cas9 system does not affect the fertility of male mice.J Reprod Immunol. 2024 Jun;163:104252. doi: 10.1016/j.jri.2024.104252. Epub 2024 Apr 29. J Reprod Immunol. 2024. PMID: 38697008
-
CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice†.Biol Reprod. 2019 Aug 1;101(2):501-511. doi: 10.1093/biolre/ioz103. Biol Reprod. 2019. PMID: 31201419 Free PMC article.
-
CRISPR-Cas9-mediated mutation revealed BSPH2 protein is dispensable for male fertility.Mol Reprod Dev. 2018 Aug;85(8-9):709-719. doi: 10.1002/mrd.23039. Epub 2018 Aug 16. Mol Reprod Dev. 2018. PMID: 29969530
-
CRISPR/Cas9-mediated genome editing in sea urchins.Methods Cell Biol. 2019;151:305-321. doi: 10.1016/bs.mcb.2018.10.004. Epub 2018 Nov 29. Methods Cell Biol. 2019. PMID: 30948015 Free PMC article. Review.
-
Disrupting the male germ line to find infertility and contraception targets.Ann Endocrinol (Paris). 2014 May;75(2):101-8. doi: 10.1016/j.ando.2014.04.006. Epub 2014 Apr 30. Ann Endocrinol (Paris). 2014. PMID: 24793995 Review.
Cited by
-
Individual disruption of 12 testis-enriched genes via the CRISPR/Cas9 system does not affect the fertility of male mice.J Reprod Immunol. 2024 Jun;163:104252. doi: 10.1016/j.jri.2024.104252. Epub 2024 Apr 29. J Reprod Immunol. 2024. PMID: 38697008
-
"Progressive motility" in elucidating novel genetic causes of male infertility.Asian J Androl. 2022 May-Jun;24(3):229-230. doi: 10.4103/aja202231. Asian J Androl. 2022. PMID: 35532567 Free PMC article. No abstract available.
-
Two acquired mouse Y chromosome-linked genes, Prssly and Teyorf1, are dispensable for male fertility‡.Biol Reprod. 2022 Sep 12;107(3):752-764. doi: 10.1093/biolre/ioac084. Biol Reprod. 2022. PMID: 35485405 Free PMC article.
-
Commentary on "CRISPR/Cas9-mediated genome editing reveals 12 testis-enriched genes dispensable for male fertility in mice".Asian J Androl. 2022 May-Jun;24(3):273. doi: 10.4103/aja202196. Asian J Androl. 2022. PMID: 34810375 Free PMC article. No abstract available.
-
Combined Use of Whole Exome Sequencing and CRISPR/Cas9 to Study the Etiology of Non-Obstructive Azoospermia: Demonstration of the Dispensable Role of the Testis-Specific Genes C1orf185 and CCT6B.Cells. 2021 Dec 30;11(1):118. doi: 10.3390/cells11010118. Cells. 2021. PMID: 35011680 Free PMC article.
References
-
- Dada R, Thilagavathi J, Venkatesh S, Esteves SC, Agarwal A. Genetic testing in male infertility. Open Reprod Sci J. 2011;3:42–56.
-
- Eddy EM, Toshimori K, O’Brien DA. Fibrous sheath of mammalian spermatozoa. Microsc Res Tech. 2003;61:103–15. - PubMed
-
- Lehti MS, Sironen A. Formation and function of sperm tail structures in association with sperm motility defects. Biol Reprod. 2017;97:522–36. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

