COVID-19: Myths and Reality
- PMID: 34284707
- PMCID: PMC8265000
- DOI: 10.1134/S0006297921070026
COVID-19: Myths and Reality
Abstract
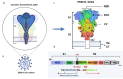
COVID-19, a new human respiratory disease that has killed nearly 3 million people in a year since the start of the pandemic, is a global public health challenge. Its infectious agent, SARS-CoV-2, differs from other coronaviruses in a number of structural features that make this virus more pathogenic and transmissible. In this review, we discuss some important characteristics of the main SARS-CoV-2 surface antigen, the spike (S) protein, such as (i) ability of the receptor-binding domain (RBD) to switch between the "standing-up" position (open pre-fusion conformation) for receptor binding and the "lying-down" position (closed pre-fusion conformation) for immune system evasion; (ii) advantage of a high binding affinity of the RBD open conformation to the human angiotensin-converting enzyme 2 (ACE2) receptor for efficient cell entry; and (iii) S protein preliminary activation by the intracellular furin-like proteases for facilitation of the virus spreading across different cell types. We describe interactions between the S protein and cellular receptors, co-receptors, and antagonists, as well as a hypothetical mechanism of the homotrimeric spike structure destabilization that triggers the fusion of the viral envelope with the cell membrane at physiological pH and mediates the viral nucleocapsid entry into the cytoplasm. The transition of the S protein pre-fusion conformation to the post-fusion one on the surface of virions after their treatment with some reagents, such as β-propiolactone, is essential, especially in relation to the vaccine production. We also compare the COVID-19 pathogenesis with that of severe outbreaks of "avian" influenza caused by the A/H5 and A/H7 highly pathogenic viruses and discuss the structural similarities between the SARS-CoV-2 S protein and hemagglutinins of those highly pathogenic strains. Finally, we touch on the prospective and currently used COVID-19 antiviral and anti-pathogenetic therapeutics, as well as recently approved conventional and innovative COVID-19 vaccines and their molecular and immunological features.
Keywords: COVID-19; S protein; SARS-CoV-2; hemagglutinin; influenza virus; structure; vaccines.
Conflict of interest statement
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
Figures



Similar articles
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
-
SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution.Nat Microbiol. 2021 Sep;6(9):1188-1198. doi: 10.1038/s41564-021-00954-4. Epub 2021 Aug 16. Nat Microbiol. 2021. PMID: 34400835
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection.J Biol Chem. 2021 Jan-Jun;296:100135. doi: 10.1074/jbc.REV120.015980. Epub 2020 Dec 6. J Biol Chem. 2021. PMID: 33268377 Free PMC article. Review.
Cited by
-
Recombinant Protein Vaccines against Human Betacoronaviruses: Strategies, Approaches and Progress.Int J Mol Sci. 2023 Jan 15;24(2):1701. doi: 10.3390/ijms24021701. Int J Mol Sci. 2023. PMID: 36675218 Free PMC article. Review.
-
Potential Coronaviral Inhibitors of the Nucleocapsid Protein Identified In Silico and In Vitro from a Large Natural Product Library.Pharmaceuticals (Basel). 2022 Aug 24;15(9):1046. doi: 10.3390/ph15091046. Pharmaceuticals (Basel). 2022. PMID: 36145267 Free PMC article.
-
From Emergence to Endemicity: A Comprehensive Review of COVID-19.Cureus. 2023 Oct 31;15(10):e48046. doi: 10.7759/cureus.48046. eCollection 2023 Oct. Cureus. 2023. PMID: 37916248 Free PMC article. Review.
-
Vascular dysfunction in COVID-19 patients: update on SARS-CoV-2 infection of endothelial cells and the role of long non-coding RNAs.Clin Sci (Lond). 2022 Nov 11;136(21):1571-1590. doi: 10.1042/CS20220235. Clin Sci (Lond). 2022. PMID: 36367091 Free PMC article.
-
Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain.Int J Mol Sci. 2023 Nov 16;24(22):16421. doi: 10.3390/ijms242216421. Int J Mol Sci. 2023. PMID: 38003610 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

