FKBP3 Induces Human Immunodeficiency Virus Type 1 Latency by Recruiting Histone Deacetylase 1/2 to the Viral Long Terminal Repeat
- PMID: 34281390
- PMCID: PMC8406261
- DOI: 10.1128/mBio.00795-21
FKBP3 Induces Human Immunodeficiency Virus Type 1 Latency by Recruiting Histone Deacetylase 1/2 to the Viral Long Terminal Repeat
Abstract
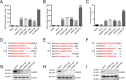
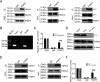
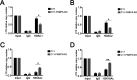
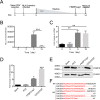
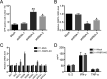
Human immunodeficiency virus type 1 (HIV-1) cannot be completely eliminated because of existence of the latent HIV-1 reservoir. However, the facts of HIV-1 latency, including its establishment and maintenance, are incomplete. FKBP3, encoded by the FKBP3 gene, belongs to the immunophilin family of proteins and is involved in immunoregulation and such cellular processes as protein folding. In a previous study, we found that FKBP3 may be related to HIV-1 latency using CRISPR screening. In this study, we knocked out the FKBP3 gene in multiple latently infected cell lines to promote latent HIV-1 activation. We found that FKBP3 could indirectly bind to the HIV-1 long terminal repeat through interaction with YY1, thereby recruiting histone deacetylase 1/2 to it. This promotes histone deacetylation and induces HIV-1 latency. Finally, in a primary latent cell model, we confirmed the effect of FKBP3 knockout on the latent activation of HIV-1. Our results suggest a new mechanism for the epigenetic regulation of HIV-1 latency and a new potential target for activating latent HIV-1. IMPORTANCE The primary reason why AIDS cannot be completely cured is the existence of a latent HIV-1 reservoir. Currently, the facts of HIV-1 latency, including its establishment and maintenance, are incomplete. Using a CRISPR library in our earlier screening of genes related to HIV-1 latency, we identified FBKP3 as a candidate gene related to HIV-1 latency. Therefore, in this mechanistic study, we first confirmed the HIV-1 latency-promoting effect of FKBP3 and determined that FKBP3 promotes histone deacetylation by recruiting histone deacetylase 1/2 to the HIV-1 long terminal repeat. We also confirmed, for the first time, that FKBP3 can act as a transcription factor (TF) recruitment scaffold and participate in epigenetic regulation of HIV-1 latency. These findings suggest a new mechanism for the epigenetic regulation of HIV-1 latency and a new potential target for activating latent HIV-1.
Keywords: FKBP3; HDAC1/2; HIV-1; HIV-1 latency; acetylation.
Figures

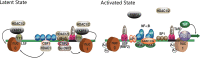
Similar articles
-
NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation.EMBO J. 2006 Jan 11;25(1):139-49. doi: 10.1038/sj.emboj.7600900. Epub 2005 Dec 1. EMBO J. 2006. PMID: 16319923 Free PMC article.
-
Inhibitors of histone deacetylases: correlation between isoform specificity and reactivation of HIV type 1 (HIV-1) from latently infected cells.J Biol Chem. 2011 Jun 24;286(25):22211-8. doi: 10.1074/jbc.M110.180224. Epub 2011 Apr 29. J Biol Chem. 2011. PMID: 21531716 Free PMC article.
-
PIWIL4 Maintains HIV-1 Latency by Enforcing Epigenetically Suppressive Modifications on the 5' Long Terminal Repeat.J Virol. 2020 May 4;94(10):e01923-19. doi: 10.1128/JVI.01923-19. Print 2020 May 4. J Virol. 2020. PMID: 32161174 Free PMC article.
-
Efficient Non-Epigenetic Activation of HIV Latency through the T-Cell Receptor Signalosome.Viruses. 2020 Aug 8;12(8):868. doi: 10.3390/v12080868. Viruses. 2020. PMID: 32784426 Free PMC article. Review.
-
Role of histone modification on transcriptional regulation and HIV-1 gene expression: possible mechanisms of periodontal diseases in AIDS progression.J Oral Sci. 2011 Mar;53(1):1-13. doi: 10.2334/josnusd.53.1. J Oral Sci. 2011. PMID: 21467809 Review.
Cited by
-
Targeting and Understanding HIV Latency: The CRISPR System against the Provirus.Pathogens. 2021 Sep 28;10(10):1257. doi: 10.3390/pathogens10101257. Pathogens. 2021. PMID: 34684206 Free PMC article. Review.
-
TRIM5α recruits HDAC1 to p50 and Sp1 and promotes H3K9 deacetylation at the HIV-1 LTR.Nat Commun. 2023 Jun 8;14(1):3343. doi: 10.1038/s41467-023-39056-6. Nat Commun. 2023. PMID: 37291137 Free PMC article.
-
Host cells reprogram lipid droplet synthesis through YY1 to resist PRRSV infection.mBio. 2024 Aug 14;15(8):e0154924. doi: 10.1128/mbio.01549-24. Epub 2024 Jul 2. mBio. 2024. PMID: 38953350 Free PMC article.
-
Proline Isomerization: From the Chemistry and Biology to Therapeutic Opportunities.Biology (Basel). 2023 Jul 14;12(7):1008. doi: 10.3390/biology12071008. Biology (Basel). 2023. PMID: 37508437 Free PMC article. Review.
-
FBXO34 promotes latent HIV-1 activation by post-transcriptional modulation.Emerg Microbes Infect. 2022 Dec;11(1):2785-2799. doi: 10.1080/22221751.2022.2140605. Emerg Microbes Infect. 2022. PMID: 36285453 Free PMC article.
References
-
- Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 96:15109–15114. doi:10.1073/pnas.96.26.15109. - DOI - PMC - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi:10.1126/science.278.5341.1295. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
