Human Beta-Defensin 2 and 3 Inhibit HIV-1 Replication in Macrophages
- PMID: 34277460
- PMCID: PMC8281893
- DOI: 10.3389/fcimb.2021.535352
Human Beta-Defensin 2 and 3 Inhibit HIV-1 Replication in Macrophages
Abstract
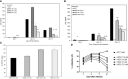
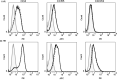
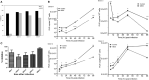
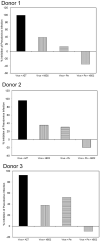
Human beta-defensins (hBDs) are broad-spectrum antimicrobial peptides, secreted by epithelial cells of the skin and mucosae, and astrocytes, which we and others have shown to inhibit HIV-1 in primary CD4+ T cells. Although loss of CD4+ T cells contributes to mucosal immune dysfunction, macrophages are a major source of persistence and spread of HIV and also contribute to the development of various HIV-associated complications. We hypothesized that, besides T cells, hBDs could protect macrophages from HIV. Our data in primary human monocyte-derived macrophages (MDM) in vitro show that hBD2 and hBD3 inhibit HIV replication in a dose-dependent manner. We determined that hBD2 neither alters surface expression of HIV receptors nor induces expression of anti-HIV cytokines or beta-chemokines in MDM. Studies using a G-protein signaling antagonist in a single-cycle reporter virus system showed that hBD2 suppresses HIV at an early post-entry stage via G-protein coupled receptor (GPCR)-mediated signaling. We find that MDM express the shared chemokine-hBD receptors CCR2 and CCR6, albeit at variable levels among donors. However, cell surface expression analyses show that neither of these receptors is necessary for hBD2-mediated HIV inhibition, suggesting that hBD2 can signal via additional receptor(s). Our data also illustrate that hBD2 treatment was associated with increased expression of APOBEC3A and 3G antiretroviral restriction factors in MDM. These findings suggest that hBD2 inhibits HIV in MDM via more than one CCR thus adding to the potential of using β-defensins in preventive and therapeutic approaches.
Keywords: APOBEC3A; APOBEC3G; CCRs; HIV-1; human β-defensin 2; macrophages.
Copyright © 2021 Bharucha, Sun, Lu, Gartner and Garzino-Demo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection.J Virol. 2005 Nov;79(22):14318-29. doi: 10.1128/JVI.79.22.14318-14329.2005. J Virol. 2005. PMID: 16254366 Free PMC article.
-
Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2.J Immunol. 2010 Jun 15;184(12):6688-94. doi: 10.4049/jimmunol.0903984. Epub 2010 May 17. J Immunol. 2010. PMID: 20483750 Free PMC article.
-
Human Beta Defensin 2 Selectively Inhibits HIV-1 in Highly Permissive CCR6⁺CD4⁺ T Cells.Viruses. 2017 May 16;9(5):111. doi: 10.3390/v9050111. Viruses. 2017. PMID: 28509877 Free PMC article.
-
Human β-defensin 2: a connection between infections and allergic skin diseases.Acta Dermatovenerol Alp Pannonica Adriat. 2024 Sep;33(3):135-139. Acta Dermatovenerol Alp Pannonica Adriat. 2024. PMID: 39324351 Review.
-
The macrophage response to HIV-1: Intracellular control of X4 virus replication accompanied by activation of chemokine and cytokine synthesis.J Neurovirol. 2002 Dec;8(6):599-610. doi: 10.1080/13550280290100923. J Neurovirol. 2002. PMID: 12476353 Review.
Cited by
-
The Papain-like Protease Domain of Severe Acute Respiratory Syndrome Coronavirus 2 Conjugated with Human Beta-Defensin 2 and Co1 Induces Mucosal and Systemic Immune Responses against the Virus.Vaccines (Basel). 2024 Apr 19;12(4):441. doi: 10.3390/vaccines12040441. Vaccines (Basel). 2024. PMID: 38675823 Free PMC article.
-
Effects of Ellagic Acid on Vaginal Innate Immune Mediators and HPV16 Infection In Vitro.Molecules. 2024 Jul 31;29(15):3630. doi: 10.3390/molecules29153630. Molecules. 2024. PMID: 39125034 Free PMC article.
-
Decreased MIP-3α Production from Antigen-Activated PBMCs in Symptomatic HIV-Infected Subjects.Pathogens. 2021 Dec 22;11(1):7. doi: 10.3390/pathogens11010007. Pathogens. 2021. PMID: 35055955 Free PMC article.
-
Defensins: defenders of human reproductive health.Hum Reprod Update. 2023 Jan 5;29(1):126-154. doi: 10.1093/humupd/dmac032. Hum Reprod Update. 2023. PMID: 36130055 Free PMC article. Review.
-
Enhancement of the activity of the antimicrobial peptides HNP1 and LL-37 by bovine pancreatic ribonuclease A.F1000Res. 2023 Mar 13;11:933. doi: 10.12688/f1000research.123044.3. eCollection 2022. F1000Res. 2023. PMID: 37360940 Free PMC article.
References
-
- Aguilar-Jimenez W., Zapata W., Caruz A., Rugeles M. T. (2013). High Transcript Levels of Vitamin D Receptor are Correlated With Higher mRNA Expression of Human Beta Defensins and IL-10 in Mucosa of HIV-1-Exposed Seronegative Individuals. PloS One 8, e82717. 10.1371/journal.pone.0082717 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

