Experimental and Genomic Evaluation of the Oestrogen Degrading Bacterium Rhodococcus equi ATCC13557
- PMID: 34276604
- PMCID: PMC8281962
- DOI: 10.3389/fmicb.2021.670928
Experimental and Genomic Evaluation of the Oestrogen Degrading Bacterium Rhodococcus equi ATCC13557
Abstract
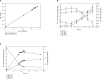
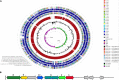
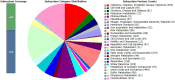
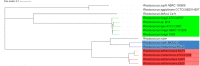
Rhodococcus equi ATCC13557 was selected as a model organism to study oestrogen degradation based on its previous ability to degrade 17α-ethinylestradiol (EE2). Biodegradation experiments revealed that R. equi ATCC13557 was unable to metabolise EE2. However, it was able to metabolise E2 with the major metabolite being E1 with no further degradation of E1. However, the conversion of E2 into E1 was incomplete, with 11.2 and 50.6% of E2 degraded in mixed (E1-E2-EE2) and E2-only conditions, respectively. Therefore, the metabolic pathway of E2 degradation by R. equi ATCC13557 may have two possible pathways. The genome of R. equi ATCC13557 was sequenced, assembled, and mapped for the first time. The genome analysis allowed the identification of genes possibly responsible for the observed biodegradation characteristics of R. equi ATCC13557. Several genes within R. equi ATCC13557 are similar, but not identical in sequence, to those identified within the genomes of other oestrogen degrading bacteria, including Pseudomonas putida strain SJTE-1 and Sphingomonas strain KC8. Homologous gene sequences coding for enzymes potentially involved in oestrogen degradation, most commonly a cytochrome P450 monooxygenase (oecB), extradiol dioxygenase (oecC), and 17β-hydroxysteroid dehydrogenase (oecA), were identified within the genome of R. equi ATCC13557. These searches also revealed a gene cluster potentially coding for enzymes involved in steroid/oestrogen degradation; 3-carboxyethylcatechol 2,3-dioxygenase, 2-hydroxymuconic semialdehyde hydrolase, 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase, 3-(3-hydroxy-phenyl)propionate hydroxylase, cytochrome P450 monooxygenase, and 3-oxosteroid 1-dehydrogenase. Further, the searches revealed steroid hormone metabolism gene clusters from the 9, 10-seco pathway, therefore R. equi ATCC13557 also has the potential to metabolise other steroid hormones such as cholesterol.
Keywords: Rhodococcus equi; bacteria; degradation; genes; genome; oestrogen.
Copyright © 2021 Harthern-Flint, Dolfing, Mrozik, Meynet, Eland, Sim and Davenport.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Mechanism of 17β-estradiol degradation by Rhodococcus equi via the 4,5-seco pathway and its key genes.Environ Pollut. 2022 Nov 1;312:120021. doi: 10.1016/j.envpol.2022.120021. Epub 2022 Aug 26. Environ Pollut. 2022. PMID: 36037852
-
Functional analysis, diversity, and distribution of the ean cluster responsible for 17β-estradiol degradation in sphingomonads.Appl Environ Microbiol. 2024 May 21;90(5):e0197423. doi: 10.1128/aem.01974-23. Epub 2024 Apr 15. Appl Environ Microbiol. 2024. PMID: 38619269 Free PMC article.
-
The Analysis of Estrogen-Degrading and Functional Metabolism Genes in Rhodococcus equi DSSKP-R-001.Int J Genomics. 2020 Aug 25;2020:9369182. doi: 10.1155/2020/9369182. eCollection 2020. Int J Genomics. 2020. PMID: 32908857 Free PMC article.
-
Pathways and genes involved in steroid hormone metabolism in male pigs: a review and update.J Steroid Biochem Mol Biol. 2014 Mar;140:44-55. doi: 10.1016/j.jsbmb.2013.11.001. Epub 2013 Nov 12. J Steroid Biochem Mol Biol. 2014. PMID: 24239507 Review.
-
Assessment of steroidogenesis and steroidogenic enzyme functions.J Steroid Biochem Mol Biol. 2013 Sep;137:176-82. doi: 10.1016/j.jsbmb.2013.05.017. Epub 2013 Jun 13. J Steroid Biochem Mol Biol. 2013. PMID: 23770321 Review.
Cited by
-
Microbial degradation of contaminants of emerging concern: metabolic, genetic and omics insights for enhanced bioremediation.Front Bioeng Biotechnol. 2024 Sep 19;12:1470522. doi: 10.3389/fbioe.2024.1470522. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39364263 Free PMC article. Review.
-
Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future.Front Microbiol. 2022 Sep 29;13:967127. doi: 10.3389/fmicb.2022.967127. eCollection 2022. Front Microbiol. 2022. PMID: 36246215 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

