Engrailed 1 coordinates cytoskeletal reorganization to induce myofibroblast differentiation
- PMID: 34259830
- PMCID: PMC8288503
- DOI: 10.1084/jem.20201916
Engrailed 1 coordinates cytoskeletal reorganization to induce myofibroblast differentiation
Abstract
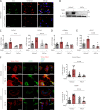
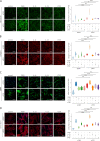
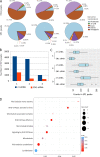
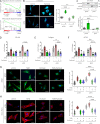
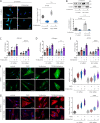
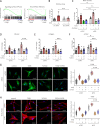
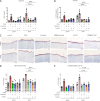
Transforming growth factor-β (TGFβ) is a key mediator of fibroblast activation in fibrotic diseases, including systemic sclerosis. Here we show that Engrailed 1 (EN1) is reexpressed in multiple fibroblast subpopulations in the skin of SSc patients. We characterize EN1 as a molecular amplifier of TGFβ signaling in myofibroblast differentiation: TGFβ induces EN1 expression in a SMAD3-dependent manner, and in turn, EN1 mediates the profibrotic effects of TGFβ. RNA sequencing demonstrates that EN1 induces a profibrotic gene expression profile functionally related to cytoskeleton organization and ROCK activation. EN1 regulates gene expression by modulating the activity of SP1 and other SP transcription factors, as confirmed by ChIP-seq experiments for EN1 and SP1. Functional experiments confirm the coordinating role of EN1 on ROCK activity and the reorganization of cytoskeleton during myofibroblast differentiation, in both standard fibroblast culture systems and in vitro skin models. Consistently, mice with fibroblast-specific knockout of En1 demonstrate impaired fibroblast-to-myofibroblast transition and are partially protected from experimental skin fibrosis.
© 2021 Györfi et al.
Conflict of interest statement
Disclosures: C. Bergmann reported personal fees from Boehringer Ingelheim and Pfizer during the conduct of the study. O. Distler reported personal fees from Abbvie, Acceleron Pharma, Amgen, AnaMar, Arxx Therapeutics, Baecon Discovery, Blade Therapeutics, Bayer, Böhringer Ingelheim, ChemomAb, Corbus Pharmacheuticals, CSL Behring, Galapagos NV, Glenmark Pharmaceuticals, GSK, Horizon Pharmaceuticals, Inventiva, Iqvia, Italfarmaco, Iqone, Kymera Therapeutics, Lupin Pharmaceuticals, Medac, Medscape, Mitsubishi Tanabe Pharma, MSD, Novartis, Pfizer, Roche, Roivant, Sanofi, Serodapharm, Topadur, Target Bioscience, and UCB during the conduct of the study; in addition, O. Distler had a patent to mir-29 for the treatment of systemic sclerosis issued (US8247389, EP2331143). J.H.W. Distler reported personal fees from Janssen, Anamar, ARXX, Bayer Pharma, Boehringer Ingelheim, Galapagos, GSK, Inventiva, Novartis, and UCB; grants from Anamar, ARXX, aTyr, Bayer Pharma, Boehringer Ingelheim, Cantargia, Celgene, CSL Behring, Galapagos, Inventiva, Kiniksa, and UCB outside the submitted work; and owns stock in 4D Science. No other disclosures were reported.
Figures



Similar articles
-
The nuclear receptor constitutive androstane receptor/NR1I3 enhances the profibrotic effects of transforming growth factor β and contributes to the development of experimental dermal fibrosis.Arthritis Rheumatol. 2014 Nov;66(11):3140-50. doi: 10.1002/art.38819. Arthritis Rheumatol. 2014. PMID: 25155144
-
Rho-associated kinases are crucial for myofibroblast differentiation and production of extracellular matrix in scleroderma fibroblasts.Arthritis Rheum. 2008 Aug;58(8):2553-64. doi: 10.1002/art.23677. Arthritis Rheum. 2008. PMID: 18668558
-
Acyltransferase skinny hedgehog regulates TGFβ-dependent fibroblast activation in SSc.Ann Rheum Dis. 2019 Sep;78(9):1269-1273. doi: 10.1136/annrheumdis-2019-215066. Epub 2019 Jun 8. Ann Rheum Dis. 2019. PMID: 31177096
-
TGFβ signaling and the control of myofibroblast differentiation: Implications for chronic inflammatory disorders.J Cell Physiol. 2018 Jan;233(1):98-106. doi: 10.1002/jcp.25879. Epub 2017 May 15. J Cell Physiol. 2018. PMID: 28247933 Review.
-
LIM-domain proteins in transforming growth factor β-induced epithelial-to-mesenchymal transition and myofibroblast differentiation.Cell Signal. 2012 Apr;24(4):819-25. doi: 10.1016/j.cellsig.2011.12.004. Epub 2011 Dec 11. Cell Signal. 2012. PMID: 22182513 Review.
Cited by
-
TGFβ3, dibutyryl cAMP and a notch inhibitor modulate phenotype late in stem cell-derived dopaminergic neuron maturation.Front Cell Dev Biol. 2023 Feb 1;11:1111705. doi: 10.3389/fcell.2023.1111705. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36819101 Free PMC article.
-
A Personalized Approach to Treat Advanced Stage Severely Contracted Joints in Dupuytren's Disease with a Unique Skeletal Distraction Device-Utilizing Modern Imaging Tools to Enhance Safety for the Patient.J Pers Med. 2022 Mar 1;12(3):378. doi: 10.3390/jpm12030378. J Pers Med. 2022. PMID: 35330378 Free PMC article.
-
Single-cell and bulk tissue sequencing unravels the heterogeneity of synovial microenvironment in arthrofibrosis.iScience. 2023 Jul 13;26(9):107379. doi: 10.1016/j.isci.2023.107379. eCollection 2023 Sep 15. iScience. 2023. PMID: 37705954 Free PMC article.
-
The Human Dermis as a Target of Nanoparticles for Treating Skin Conditions.Pharmaceutics. 2022 Dec 20;15(1):10. doi: 10.3390/pharmaceutics15010010. Pharmaceutics. 2022. PMID: 36678639 Free PMC article. Review.
-
CD201+ fascia progenitors choreograph injury repair.Nature. 2023 Nov;623(7988):792-802. doi: 10.1038/s41586-023-06725-x. Epub 2023 Nov 15. Nature. 2023. PMID: 37968392 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

