Protein conformational switch discerned via network centrality properties
- PMID: 34257839
- PMCID: PMC8246261
- DOI: 10.1016/j.csbj.2021.06.004
Protein conformational switch discerned via network centrality properties
Abstract
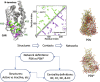
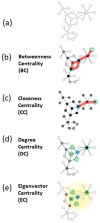
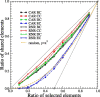
Network analysis has emerged as a powerful tool for examining structural biology systems. The spatial organization of the components of a biomolecular structure has been rendered as a graph representation and analyses have been performed to deduce the biophysical and mechanistic properties of these components. For proteins, the analysis of protein structure networks (PSNs), especially via network centrality measurements and cluster coefficients, has led to identifying amino acid residues that play key functional roles and classifying amino acid residues in general. Whether these network properties examined in various studies are sensitive to subtle (yet biologically significant) conformational changes remained to be addressed. Here, we focused on four types of network centrality properties (betweenness, closeness, degree, and eigenvector centralities) for conformational changes upon ligand binding of a sensor protein (constitutive androstane receptor) and an allosteric enzyme (ribonucleotide reductase). We found that eigenvector centrality is sensitive and can distinguish salient structural features between protein conformational states while other centrality measures, especially closeness centrality, are less sensitive and rather generic with respect to the structural specificity. We also demonstrated that an ensemble-informed, modified PSN with static edges removed (which we term PSN*) has enhanced sensitivity at discerning structural changes.
Keywords: Conformational switch; Network analysis; Network centrality; Protein structure network.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
An Analysis of Central Residues Between Ligand-Bound and Ligand-Free Protein Structures Based on Network Approach.Protein Pept Lett. 2017 Aug;24(6):517-527. doi: 10.2174/0929866524666170413120940. Protein Pept Lett. 2017. PMID: 28412901
-
Centrality Measures in Residue Interaction Networks to Highlight Amino Acids in Protein-Protein Binding.Front Bioinform. 2021 Jun 18;1:684970. doi: 10.3389/fbinf.2021.684970. eCollection 2021. Front Bioinform. 2021. PMID: 36303777 Free PMC article.
-
Centralities in simplicial complexes. Applications to protein interaction networks.J Theor Biol. 2018 Feb 7;438:46-60. doi: 10.1016/j.jtbi.2017.11.003. Epub 2017 Nov 8. J Theor Biol. 2018. PMID: 29128505
-
What do centrality measures measure in psychological networks?J Abnorm Psychol. 2019 Nov;128(8):892-903. doi: 10.1037/abn0000446. Epub 2019 Jul 18. J Abnorm Psychol. 2019. PMID: 31318245 Review.
-
Intra and inter-molecular communications through protein structure network.Curr Protein Pept Sci. 2009 Apr;10(2):146-60. doi: 10.2174/138920309787847590. Curr Protein Pept Sci. 2009. PMID: 19355982 Review.
Cited by
-
Understanding the P-Loop Conformation in the Determination of Inhibitor Selectivity Toward the Hepatocellular Carcinoma-Associated Dark Kinase STK17B.Front Mol Biosci. 2022 May 10;9:901603. doi: 10.3389/fmolb.2022.901603. eCollection 2022. Front Mol Biosci. 2022. PMID: 35620482 Free PMC article.
-
Rinmaker: a fast, versatile and reliable tool to determine residue interaction networks in proteins.BMC Bioinformatics. 2023 Sep 11;24(1):336. doi: 10.1186/s12859-023-05466-y. BMC Bioinformatics. 2023. PMID: 37697267 Free PMC article.
-
The "violin model": Looking at community networks for dynamic allostery.J Chem Phys. 2023 Feb 28;158(8):081001. doi: 10.1063/5.0138175. J Chem Phys. 2023. PMID: 36859094 Free PMC article. Review.
-
Identifying Tumor-Associated Genes from Bilayer Networks of DNA Methylation Sites and RNAs.Life (Basel). 2022 Dec 27;13(1):76. doi: 10.3390/life13010076. Life (Basel). 2022. PMID: 36676027 Free PMC article.
-
Autopromotion of K-Ras4B Feedback Activation Through an SOS-Mediated Long-Range Allosteric Effect.Front Mol Biosci. 2022 Apr 8;9:860962. doi: 10.3389/fmolb.2022.860962. eCollection 2022. Front Mol Biosci. 2022. PMID: 35463958 Free PMC article.
References
-
- Fersht A. W.H. Freeman; New York: 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding.
-
- Jackson M.B. 1st edn. Cambridge University Press; 2006. Molecular and cellular biophysics.
-
- C.-I. Branden, J. Tooze, Introduction to protein structure, Garland Pub, New York, ISBN 9780815303442 9780815302704 9780815304869, 1991.
-
- Di Paola L., Giuliani A. Protein contact network topology: a natural language for allostery. Curr Opin Struct Biol. 2015;31:43–48. - PubMed
LinkOut - more resources
Full Text Sources
