Sphingolipids mediate polar sorting of PIN2 through phosphoinositide consumption at the trans-Golgi network
- PMID: 34257291
- PMCID: PMC8277843
- DOI: 10.1038/s41467-021-24548-0
Sphingolipids mediate polar sorting of PIN2 through phosphoinositide consumption at the trans-Golgi network
Abstract
The lipid composition of organelles acts as a landmark to define membrane identity and specify subcellular function. Phosphoinositides are anionic lipids acting in protein sorting and trafficking at the trans-Golgi network (TGN). In animal cells, sphingolipids control the turnover of phosphoinositides through lipid exchange mechanisms at endoplasmic reticulum/TGN contact sites. In this study, we discover a mechanism for how sphingolipids mediate phosphoinositide homeostasis at the TGN in plant cells. Using multiple approaches, we show that a reduction of the acyl-chain length of sphingolipids results in an increased level of phosphatidylinositol-4-phosphate (PtdIns(4)P or PI4P) at the TGN but not of other lipids usually coupled to PI4P during exchange mechanisms. We show that sphingolipids mediate Phospholipase C (PLC)-driven consumption of PI4P at the TGN rather than local PI4P synthesis and that this mechanism is involved in the polar sorting of the auxin efflux carrier PIN2 at the TGN. Together, our data identify a mode of action of sphingolipids in lipid interplay at the TGN during protein sorting.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
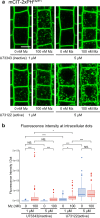
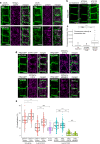
Similar articles
-
Sphingolipid metabolic flow controls phosphoinositide turnover at the trans-Golgi network.EMBO J. 2017 Jun 14;36(12):1736-1754. doi: 10.15252/embj.201696048. Epub 2017 May 10. EMBO J. 2017. PMID: 28495678 Free PMC article.
-
Enrichment of hydroxylated C24- and C26-acyl-chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains.Nat Commun. 2016 Sep 29;7:12788. doi: 10.1038/ncomms12788. Nat Commun. 2016. PMID: 27681606 Free PMC article.
-
Phosphoinositides and membrane traffic at the trans-Golgi network.Biochem Soc Symp. 2005;(72):31-8. doi: 10.1042/bss0720031. Biochem Soc Symp. 2005. PMID: 15649127 Review.
-
Illuminating the membrane contact sites between the endoplasmic reticulum and the trans-Golgi network.FEBS Lett. 2019 Nov;593(22):3135-3148. doi: 10.1002/1873-3468.13639. Epub 2019 Nov 9. FEBS Lett. 2019. PMID: 31610025 Review.
-
Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network.J Cell Biol. 2009 May 18;185(4):601-12. doi: 10.1083/jcb.200901145. Epub 2009 May 11. J Cell Biol. 2009. PMID: 19433450 Free PMC article.
Cited by
-
Subcellular trafficking and post-translational modification regulate PIN polarity in plants.Front Plant Sci. 2022 Jul 27;13:923293. doi: 10.3389/fpls.2022.923293. eCollection 2022. Front Plant Sci. 2022. PMID: 35968084 Free PMC article. Review.
-
Inhibition of Very Long Chain Fatty Acids Synthesis Mediates PI3P Homeostasis at Endosomal Compartments.Int J Mol Sci. 2021 Aug 6;22(16):8450. doi: 10.3390/ijms22168450. Int J Mol Sci. 2021. PMID: 34445155 Free PMC article.
-
Lipid Polarization during Cytokinesis.Cells. 2022 Dec 8;11(24):3977. doi: 10.3390/cells11243977. Cells. 2022. PMID: 36552741 Free PMC article. Review.
-
ER Membrane Lipid Composition and Metabolism: Lipidomic Analysis.Methods Mol Biol. 2024;2772:137-148. doi: 10.1007/978-1-0716-3710-4_10. Methods Mol Biol. 2024. PMID: 38411811
-
StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana.Viruses. 2022 Mar 10;14(3):574. doi: 10.3390/v14030574. Viruses. 2022. PMID: 35336981 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

