Treponema denticola dentilisin triggered TLR2/MyD88 activation upregulates a tissue destructive program involving MMPs via Sp1 in human oral cells
- PMID: 34255809
- PMCID: PMC8301614
- DOI: 10.1371/journal.ppat.1009311
Treponema denticola dentilisin triggered TLR2/MyD88 activation upregulates a tissue destructive program involving MMPs via Sp1 in human oral cells
Abstract
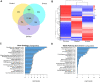
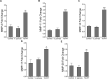
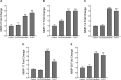
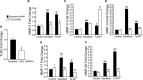
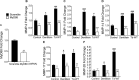
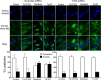
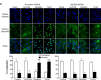
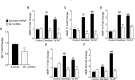
Periodontal disease is driven by dysbiosis in the oral microbiome, resulting in over-representation of species that induce the release of pro-inflammatory cytokines, chemokines, and tissue-remodeling matrix metalloproteinases (MMPs) in the periodontium. These chronic tissue-destructive inflammatory responses result in gradual loss of tooth-supporting alveolar bone. The oral spirochete Treponema denticola, is consistently found at significantly elevated levels in periodontal lesions. Host-expressed Toll-Like Receptor 2 (TLR2) senses a variety of bacterial ligands, including acylated lipopolysaccharides and lipoproteins. T. denticola dentilisin, a surface-expressed protease complex comprised of three lipoproteins has been implicated as a virulence factor in periodontal disease, primarily due to its proteolytic activity. While the role of acylated bacterial components in induction of inflammation is well-studied, little attention has been given to the potential role of the acylated nature of dentilisin. The purpose of this study was to test the hypothesis that T. denticola dentilisin activates a TLR2-dependent mechanism, leading to upregulation of tissue-destructive genes in periodontal tissue. RNA-sequencing of periodontal ligament cells challenged with T. denticola bacteria revealed significant upregulation of genes associated with extracellular matrix organization and degradation including potentially tissue-specific inducible MMPs that may play novel roles in modulating host immune responses that have yet to be characterized within the context of oral disease. The Gram-negative oral commensal, Veillonella parvula, failed to upregulate these same MMPs. Dentilisin-induced upregulation of MMPs was mediated via TLR2 and MyD88 activation, since knockdown of expression of either abrogated these effects. Challenge with purified dentilisin upregulated the same MMPs while a dentilisin-deficient T. denticola mutant had no effect. Finally, T. denticola-mediated activation of TLR2/MyD88 lead to the nuclear translocation of the transcription factor Sp1, which was shown to be a critical regulator of all T. denticola-dependent MMP expression. Taken together, these data suggest that T. denticola dentilisin stimulates tissue-destructive cellular processes in a TLR2/MyD88/Sp1-dependent fashion.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
The Treponema denticola chymotrypsin-like protease dentilisin induces matrix metalloproteinase-2-dependent fibronectin fragmentation in periodontal ligament cells.Infect Immun. 2011 Feb;79(2):806-11. doi: 10.1128/IAI.01001-10. Epub 2010 Nov 29. Infect Immun. 2011. PMID: 21115719 Free PMC article.
-
Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography.Bio Protoc. 2024 Apr 5;14(7):e4970. doi: 10.21769/BioProtoc.4970. eCollection 2024 Apr 5. Bio Protoc. 2024. PMID: 38618176 Free PMC article.
-
Stimulation of Human Periodontal Ligament Fibroblasts Using Purified Dentilisin Extracted from Treponema denticola.Bio Protoc. 2022 Dec 20;12(24):e4571. doi: 10.21769/BioProtoc.4571. eCollection 2022 Dec 20. Bio Protoc. 2022. PMID: 36618097 Free PMC article.
-
Approaches to Understanding Mechanisms of Dentilisin Protease Complex Expression in Treponema denticola.Front Cell Infect Microbiol. 2021 May 18;11:668287. doi: 10.3389/fcimb.2021.668287. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34084756 Free PMC article. Review.
-
Role of Treponema denticola in periodontal diseases.Crit Rev Oral Biol Med. 2001;12(5):399-413. doi: 10.1177/10454411010120050301. Crit Rev Oral Biol Med. 2001. PMID: 12002822 Review.
Cited by
-
Molecular docking analysis of a virulence factor protein dentilisin from Treponema denticola with oxazole piperazine derivatives.Bioinformation. 2023 Jan 31;19(1):57-62. doi: 10.6026/97320630019057. eCollection 2023. Bioinformation. 2023. PMID: 37720272 Free PMC article.
-
Illuminating the oral microbiome: cellular microbiology.FEMS Microbiol Rev. 2023 Jul 5;47(4):fuad045. doi: 10.1093/femsre/fuad045. FEMS Microbiol Rev. 2023. PMID: 37533213 Free PMC article.
-
Nisin probiotic prevents inflammatory bone loss while promoting reparative proliferation and a healthy microbiome.NPJ Biofilms Microbiomes. 2022 Jun 7;8(1):45. doi: 10.1038/s41522-022-00307-x. NPJ Biofilms Microbiomes. 2022. PMID: 35672331 Free PMC article.
-
The Emerging Role of MMP12 in the Oral Environment.Int J Mol Sci. 2023 Feb 28;24(5):4648. doi: 10.3390/ijms24054648. Int J Mol Sci. 2023. PMID: 36902078 Free PMC article. Review.
-
Nisin a probiotic bacteriocin mitigates brain microbiome dysbiosis and Alzheimer's disease-like neuroinflammation triggered by periodontal disease.J Neuroinflammation. 2023 Oct 6;20(1):228. doi: 10.1186/s12974-023-02915-6. J Neuroinflammation. 2023. PMID: 37803465 Free PMC article.
References
-
- Madianos P.N., Bobetsis Y.A., and Offenbacher S., Adverse pregnancy outcomes (APOs) and periodontal disease: pathogenic mechanisms. Journal of Periodontology, 2013. 84(4S): p. S170–S180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

