Germ cells: ENCODE's forgotten cell type†
- PMID: 34250539
- PMCID: PMC8444701
- DOI: 10.1093/biolre/ioab135
Germ cells: ENCODE's forgotten cell type†
Abstract
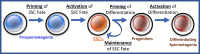
More than a decade ago, the ENCODE and NIH Epigenomics Roadmap consortia organized large multilaboratory efforts to profile the epigenomes of >110 different mammalian somatic cell types. This generated valuable publicly accessible datasets that are being mined to reveal genome-wide patterns of a variety of different epigenetic parameters. This consortia approach facilitated the powerful and comprehensive multiparametric integrative analysis of the epigenomes in each cell type. However, no germ cell types were included among the cell types characterized by either of these consortia. Thus, comprehensive epigenetic profiling data are not generally available for the most evolutionarily important cells, male and female germ cells. We discuss the need for reproductive biologists to generate similar multiparametric epigenomic profiling datasets for both male and female germ cells at different developmental stages and summarize our recent effort to derive such data for mammalian spermatogonial stem cells and progenitor spermatogonia.
Keywords: cell fate; chromatin; epigenetic profiling; progenitor spermatogonia; spermatogonial stem cells.
© The Author(s) 2021. Published by Oxford University Press on behalf of Society for the Study of Reproduction. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Spermatogonial stem cells: updates from specification to clinical relevance.Hum Reprod Update. 2019 May 1;25(3):275-297. doi: 10.1093/humupd/dmz006. Hum Reprod Update. 2019. PMID: 30810745 Review.
-
Stage-specific embryonic antigen 4 is a membrane marker for enrichment of porcine spermatogonial stem cells.Andrology. 2020 Nov;8(6):1923-1934. doi: 10.1111/andr.12870. Epub 2020 Aug 9. Andrology. 2020. PMID: 32691968
-
Epigenetic profiles signify cell fate plasticity in unipotent spermatogonial stem and progenitor cells.Nat Commun. 2016 Apr 27;7:11275. doi: 10.1038/ncomms11275. Nat Commun. 2016. PMID: 27117588 Free PMC article.
-
Integrative epigenomic analysis reveals unique epigenetic signatures involved in unipotency of mouse female germline stem cells.Genome Biol. 2016 Jul 27;17(1):162. doi: 10.1186/s13059-016-1023-z. Genome Biol. 2016. PMID: 27465593 Free PMC article.
-
Epigenetic priming in the male germline.Curr Opin Genet Dev. 2024 Jun;86:102190. doi: 10.1016/j.gde.2024.102190. Epub 2024 Apr 11. Curr Opin Genet Dev. 2024. PMID: 38608568 Free PMC article. Review.
References
-
- Stewart TA, Bellvé AR, Leder P. Transcription and promoter usage of the Myc gene in normal somatic and spermatogenic cells. Science 1984; 226:707–710. - PubMed