RAVE and Rabconnectin-3 Complexes as Signal Dependent Regulators of Organelle Acidification
- PMID: 34249946
- PMCID: PMC8264551
- DOI: 10.3389/fcell.2021.698190
RAVE and Rabconnectin-3 Complexes as Signal Dependent Regulators of Organelle Acidification
Abstract
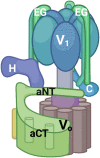
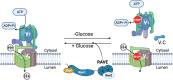
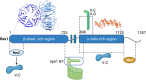
The yeast RAVE (Regulator of H+-ATPase of Vacuolar and Endosomal membranes) complex and Rabconnectin-3 complexes of higher eukaryotes regulate acidification of organelles such as lysosomes and endosomes by catalyzing V-ATPase assembly. V-ATPases are highly conserved proton pumps consisting of a peripheral V1 subcomplex that contains the sites of ATP hydrolysis, attached to an integral membrane V o subcomplex that forms the transmembrane proton pore. Reversible disassembly of the V-ATPase is a conserved regulatory mechanism that occurs in response to multiple signals, serving to tune ATPase activity and compartment acidification to changing extracellular conditions. Signals such as glucose deprivation can induce release of V1 from Vo, which inhibits both ATPase activity and proton transport. Reassembly of V1 with Vo restores ATP-driven proton transport, but requires assistance of the RAVE or Rabconnectin-3 complexes. Glucose deprivation triggers V-ATPase disassembly in yeast and is accompanied by binding of RAVE to V1 subcomplexes. Upon glucose readdition, RAVE catalyzes both recruitment of V1 to the vacuolar membrane and its reassembly with Vo. The RAVE complex can be recruited to the vacuolar membrane by glucose in the absence of V1 subunits, indicating that the interaction between RAVE and the Vo membrane domain is glucose-sensitive. Yeast RAVE complexes also distinguish between organelle-specific isoforms of the Vo a-subunit and thus regulate distinct V-ATPase subpopulations. Rabconnectin-3 complexes in higher eukaryotes appear to be functionally equivalent to yeast RAVE. Originally isolated as a two-subunit complex from rat brain, the Rabconnectin-3 complex has regions of homology with yeast RAVE and was shown to interact with V-ATPase subunits and promote endosomal acidification. Current understanding of the structure and function of RAVE and Rabconnectin-3 complexes, their interactions with the V-ATPase, their role in signal-dependent modulation of organelle acidification, and their impact on downstream pathways will be discussed.
Keywords: DMXL2; RAVE = regulator of H+-ATPase of vacuoles and endosomes; Rabconnectin-3; V-ATPase; WDR7; endosome and lysosome; organelle acidification; vacuole.
Copyright © 2021 Jaskolka, Winkley and Kane.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Interaction between the yeast RAVE complex and Vph1-containing Vo sectors is a central glucose-sensitive interaction required for V-ATPase reassembly.J Biol Chem. 2020 Feb 21;295(8):2259-2269. doi: 10.1074/jbc.RA119.011522. Epub 2020 Jan 15. J Biol Chem. 2020. PMID: 31941791 Free PMC article.
-
Molecular Interactions and Cellular Itinerary of the Yeast RAVE (Regulator of the H+-ATPase of Vacuolar and Endosomal Membranes) Complex.J Biol Chem. 2015 Nov 13;290(46):27511-23. doi: 10.1074/jbc.M115.667634. Epub 2015 Sep 24. J Biol Chem. 2015. PMID: 26405040 Free PMC article.
-
Defining steps in RAVE-catalyzed V-ATPase assembly using purified RAVE and V-ATPase subcomplexes.J Biol Chem. 2021 Jan-Jun;296:100703. doi: 10.1016/j.jbc.2021.100703. Epub 2021 Apr 22. J Biol Chem. 2021. PMID: 33895134 Free PMC article.
-
Targeting reversible disassembly as a mechanism of controlling V-ATPase activity.Curr Protein Pept Sci. 2012 Mar;13(2):117-23. doi: 10.2174/138920312800493142. Curr Protein Pept Sci. 2012. PMID: 22044153 Free PMC article. Review.
-
The cytosolic N-terminal domain of V-ATPase a-subunits is a regulatory hub targeted by multiple signals.Front Mol Biosci. 2023 Jun 16;10:1168680. doi: 10.3389/fmolb.2023.1168680. eCollection 2023. Front Mol Biosci. 2023. PMID: 37398550 Free PMC article. Review.
Cited by
-
Role of Lysosomal Acidification Dysfunction in Mesenchymal Stem Cell Senescence.Front Cell Dev Biol. 2022 Feb 7;10:817877. doi: 10.3389/fcell.2022.817877. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35198560 Free PMC article. Review.
-
Direct control of lysosomal catabolic activity by mTORC1 through regulation of V-ATPase assembly.Nat Commun. 2022 Aug 17;13(1):4848. doi: 10.1038/s41467-022-32515-6. Nat Commun. 2022. PMID: 35977928 Free PMC article.
-
Endolysosomal trafficking controls yolk granule biogenesis in vitellogenic Drosophila oocytes.PLoS Genet. 2024 Feb 5;20(2):e1011152. doi: 10.1371/journal.pgen.1011152. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38315726 Free PMC article.
-
Reversible assembly and disassembly of V-ATPase during the lysosome regeneration cycle.Mol Biol Cell. 2024 May 1;35(5):ar63. doi: 10.1091/mbc.E23-08-0322. Epub 2024 Mar 6. Mol Biol Cell. 2024. PMID: 38446621 Free PMC article.
-
Ion Channels and Pumps in Autophagy: A Reciprocal Relationship.Cells. 2021 Dec 14;10(12):3537. doi: 10.3390/cells10123537. Cells. 2021. PMID: 34944044 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

