Effect of Transgenic Rootstock Grafting on the Omics Profiles in Tomato
- PMID: 34249588
- PMCID: PMC8254850
- DOI: 10.14252/foodsafetyfscj.D-20-00032
Effect of Transgenic Rootstock Grafting on the Omics Profiles in Tomato
Abstract
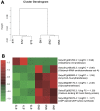
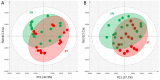
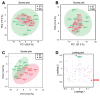
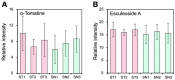
Grafting of non-transgenic scion onto genetically modified (GM) rootstocks provides superior agronomic traits in the GM rootstock, and excellent fruits can be produced for consumption. In such grafted plants, the scion does not contain any foreign genes, but the fruit itself is likely to be influenced directly or indirectly by the foreign genes in the rootstock. Before market release of such fruit products, the effects of grafting onto GM rootstocks should be determined from the perspective of safety use. Here, we evaluated the effects of a transgene encoding β-glucuronidase (GUS) on the grafted tomato fruits as a model case. An edible tomato cultivar, Stella Mini Tomato, was grafted onto GM Micro-Tom tomato plants that had been transformed with the GUS gene. The grafted plants showed no difference in their fruit development rate and fresh weight regardless of the presence or absence of the GUS gene in the rootstock. The fruit samples were subjected to transcriptome (NGS-illumina), proteome (shotgun LC-MS/MS), metabolome (LC-ESI-MS and GC-EI-MS), and general food ingredient analyses. In addition, differentially detected items were identified between the grafted plants onto rootstocks with or without transgenes (more than two-fold). The transcriptome analysis detected approximately 18,500 expressed genes on average, and only 6 genes were identified as differentially expressed. Principal component analysis of 2,442 peaks for peptides in proteome profiles showed no significant differences. In the LC-ESI-MS and GC-EI-MS analyses, a total of 93 peak groups and 114 peak groups were identified, respectively, and only 2 peak groups showed more than two-fold differences. The general food ingredient analysis showed no significant differences in the fruits of Stella scions between GM and non-GM Micro-Tom rootstocks. These multiple omics data showed that grafting on the rootstock harboring the GUS transgene did not induce any genetic or metabolic variation in the scion.
Keywords: Solanum lycopersicum; genetically modified (GM) plants; grafting; new plant breeding technology (NPBT); omics analysis; tomato.
©2021 Food Safety Commission, Cabinet Office, Government of Japan.
Conflict of interest statement
Conflict of Interest: The authors have no conflict of interest.
Figures

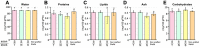
Similar articles
-
Omics Profiles of Non-transgenic Scion Grafted on Transgenic RdDM Rootstock.Food Saf (Tokyo). 2022 Mar 4;10(1):13-31. doi: 10.14252/foodsafetyfscj.D-21-00012. eCollection 2022 Mar. Food Saf (Tokyo). 2022. PMID: 35510071 Free PMC article.
-
Multi-omics Analyses of Non-GM Tomato Scion Engrafted on GM Rootstocks.Food Saf (Tokyo). 2023 Sep 6;11(3):41-53. doi: 10.14252/foodsafetyfscj.D-23-00005. eCollection 2023 Sep. Food Saf (Tokyo). 2023. PMID: 37745161 Free PMC article.
-
Modification of the Sensory Profile and Volatile Aroma Compounds of Tomato Fruits by the Scion × Rootstock Interactive Effect.Front Plant Sci. 2021 Jan 20;11:616431. doi: 10.3389/fpls.2020.616431. eCollection 2020. Front Plant Sci. 2021. PMID: 33552108 Free PMC article.
-
Vegetable Grafting From a Molecular Point of View: The Involvement of Epigenetics in Rootstock-Scion Interactions.Front Plant Sci. 2021 Jan 7;11:621999. doi: 10.3389/fpls.2020.621999. eCollection 2020. Front Plant Sci. 2021. PMID: 33488662 Free PMC article. Review.
-
Grafting: A Technique to Modify Ion Accumulation in Horticultural Crops.Front Plant Sci. 2016 Oct 21;7:1457. doi: 10.3389/fpls.2016.01457. eCollection 2016. Front Plant Sci. 2016. PMID: 27818663 Free PMC article. Review.
Cited by
-
Omics Profiles of Non-transgenic Scion Grafted on Transgenic RdDM Rootstock.Food Saf (Tokyo). 2022 Mar 4;10(1):13-31. doi: 10.14252/foodsafetyfscj.D-21-00012. eCollection 2022 Mar. Food Saf (Tokyo). 2022. PMID: 35510071 Free PMC article.
-
Impact of Intron and Retransformation on Transgene Expression in Leaf and Fruit Tissues of Field-Grown Pear Trees.Int J Mol Sci. 2023 Aug 17;24(16):12883. doi: 10.3390/ijms241612883. Int J Mol Sci. 2023. PMID: 37629068 Free PMC article.
-
Discontinuous Translocation of a Luciferase Protein beyond Graft Junction in Tobacco.Food Saf (Tokyo). 2024 Mar 22;12(1):1-16. doi: 10.14252/foodsafetyfscj.D-23-00010. eCollection 2024 Mar. Food Saf (Tokyo). 2024. PMID: 38532775 Free PMC article.
-
Recent applications of metabolomics in plant breeding.Breed Sci. 2022 Mar;72(1):56-65. doi: 10.1270/jsbbs.21065. Epub 2022 Feb 3. Breed Sci. 2022. PMID: 36045891 Free PMC article.
-
Multi-omics Analyses of Non-GM Tomato Scion Engrafted on GM Rootstocks.Food Saf (Tokyo). 2023 Sep 6;11(3):41-53. doi: 10.14252/foodsafetyfscj.D-23-00005. eCollection 2023 Sep. Food Saf (Tokyo). 2023. PMID: 37745161 Free PMC article.
References
-
- Codex Alimentarius Commission (CAC), Joint FAO/WHO Food Standards Program. Principles for the risk analysis of foods derived from modern biotechnology (CAC/GL 44-2003). 2003. http://www.fao.org/fileadmin/user_upload/gmfp/resources/ CXG_044e.pdf. Accessed on November 26, 2020.
-
- Codex Alimentarius Commission (CAC), Joint FAO/WHO Food Standards Program. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants (CAC/GL 45-2003) 2003. http://www.fao.org/fileadmin/user_upload/gmfp/docs/CAC.GL_45_2003.pdf. Accessed onNovember 26, 2020
-
- Codex Alimentarius Commission (CAC), Joint FAO/WHO Food Standards Program. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA microorganisms (CAC/GL 46-2003). 2003. http://www.fao.org/fileadmin/user_upload/gmfp/resources/CXG_046e.pdf. Accessed on November 26, 2020.
-
- Codex Alimentarius Commission (CAC), Joint FAO/WHO Food Standards Program. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA animals (CAC/GL 68-2008). 2003. http://www.fao.org/fileadmin/user_upload/gmfp/resources/CXG_068e.pdf. Accessed on November 26, 2020.
-
- Food Safety Commission of Japan. Standards for the safety assessment of genetically modified foods (seed plants). 2004. http://www.fsc.go.jp/senmon/idensi/gm_kijun.pdf. Accessed on November 26, 2020.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

