Identification of small molecules that interfere with c-di-GMP signaling and induce dispersal of Pseudomonas aeruginosa biofilms
- PMID: 34244523
- PMCID: PMC8271024
- DOI: 10.1038/s41522-021-00225-4
Identification of small molecules that interfere with c-di-GMP signaling and induce dispersal of Pseudomonas aeruginosa biofilms
Abstract
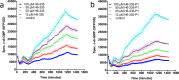
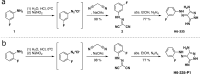
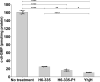
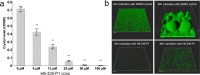
Microbial biofilms are involved in a number of infections that cannot be cured, as microbes in biofilms resist host immune defenses and antibiotic therapies. With no strict biofilm-antibiotic in the current pipelines, there is an unmet need for drug candidates that enable the current antibiotics to eradicate bacteria in biofilms. We used high-throughput screening to identify chemical compounds that reduce the intracellular c-di-GMP content in Pseudomonas aeruginosa. This led to the identification of a small molecule that efficiently depletes P. aeruginosa for c-di-GMP, inhibits biofilm formation, and disperses established biofilm. A combination of our lead compound with standard of care antibiotics showed improved eradication of an implant-associated infection established in mice. Genetic analyses provided evidence that the anti-biofilm compound stimulates the activity of the c-di-GMP phosphodiesterase BifA in P. aeruginosa. Our work constitutes a proof of concept for c-di-GMP phosphodiesterase-activating drugs administered in combination with antibiotics as a viable treatment strategy for otherwise recalcitrant infections.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests. The anti-biofilm activity of the H6-335 and H6-335-P1 compounds is protected with a patent application filed (priority date) 20.10.2020 - application no. 20193050.0-1110.
Figures


Similar articles
-
The anti-cancerous drug doxorubicin decreases the c-di-GMP content in Pseudomonas aeruginosa but promotes biofilm formation.Microbiology (Reading). 2016 Oct;162(10):1797-1807. doi: 10.1099/mic.0.000354. Epub 2016 Aug 15. Microbiology (Reading). 2016. PMID: 27526691
-
Genetic Tools to Study c-di-GMP-Dependent Signaling in Pseudomonas aeruginosa.Methods Mol Biol. 2017;1657:471-480. doi: 10.1007/978-1-4939-7240-1_34. Methods Mol Biol. 2017. PMID: 28889314
-
Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic Di-GMP level in the bacteria.Infect Immun. 2013 Aug;81(8):2705-13. doi: 10.1128/IAI.00332-13. Epub 2013 May 20. Infect Immun. 2013. PMID: 23690403 Free PMC article.
-
Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria.J Biol Chem. 2016 Jun 10;291(24):12547-12555. doi: 10.1074/jbc.R115.711507. Epub 2016 Apr 21. J Biol Chem. 2016. PMID: 27129226 Free PMC article. Review.
-
c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review.Microbiol Spectr. 2015 Apr;3(2):MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. Microbiol Spectr. 2015. PMID: 26104694 Free PMC article. Review.
Cited by
-
The dynamics of biofilm development and dispersal should be taken into account when quantifying biofilm via the crystal violet microtiter plate assay.Biofilm. 2024 Jun 20;8:100207. doi: 10.1016/j.bioflm.2024.100207. eCollection 2024 Dec. Biofilm. 2024. PMID: 39021701 Free PMC article.
-
Phenotypic and integrated analysis of a comprehensive Pseudomonas aeruginosa PAO1 library of mutants lacking cyclic-di-GMP-related genes.Front Microbiol. 2022 Jul 22;13:949597. doi: 10.3389/fmicb.2022.949597. eCollection 2022. Front Microbiol. 2022. PMID: 35935233 Free PMC article.
-
SAR study of 4-arylazo-3,5-diamino-1H-pyrazoles: identification of small molecules that induce dispersal of Pseudomonas aeruginosa biofilms.RSC Med Chem. 2021 Sep 3;12(11):1868-1878. doi: 10.1039/d1md00275a. eCollection 2021 Nov 17. RSC Med Chem. 2021. PMID: 34841247 Free PMC article.
-
In vivo evolution of antimicrobial resistance in a biofilm model of Pseudomonas aeruginosa lung infection.ISME J. 2024 Jan 8;18(1):wrae036. doi: 10.1093/ismejo/wrae036. ISME J. 2024. PMID: 38478426 Free PMC article.
-
Surfaces modified with small molecules that interfere with nucleotide signaling reduce Staphylococcus epidermidis biofilm and increase the efficacy of ciprofloxacin.Colloids Surf B Biointerfaces. 2023 Jul;227:113345. doi: 10.1016/j.colsurfb.2023.113345. Epub 2023 May 12. Colloids Surf B Biointerfaces. 2023. PMID: 37196462 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

