Recruitment of endoplasmic reticulum-targeted and cytosolic mRNAs into membrane-associated stress granules
- PMID: 34244458
- PMCID: PMC8456999
- DOI: 10.1261/rna.078858.121
Recruitment of endoplasmic reticulum-targeted and cytosolic mRNAs into membrane-associated stress granules
Abstract
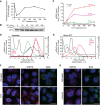
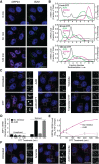
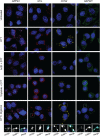
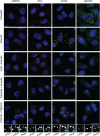
Stress granules (SGs) are membraneless organelles composed of mRNAs and RNA binding proteins which undergo assembly in response to stress-induced inactivation of translation initiation. In general, SG recruitment is limited to a subpopulation of a given mRNA species and RNA-seq analyses of purified SGs revealed that signal sequence-encoding (i.e., endoplasmic reticulum [ER]-targeted) transcripts are significantly underrepresented, consistent with prior reports that ER localization can protect mRNAs from SG recruitment. Using translational profiling, cell fractionation, and single molecule mRNA imaging, we examined SG biogenesis following activation of the unfolded protein response (UPR) by 1,4-dithiothreitol (DTT) and report that gene-specific subsets of cytosolic and ER-targeted mRNAs can be recruited into SGs. Furthermore, we demonstrate that SGs form in close proximity to or directly associated with the ER membrane. ER-associated SG assembly was also observed during arsenite stress, suggesting broad roles for the ER in SG biogenesis. Recruitment of a given mRNA into SGs required stress-induced translational repression, though translational inhibition was not solely predictive of an mRNA's propensity for SG recruitment. SG formation was prevented by the transcriptional inhibitors actinomycin D or triptolide, suggesting a functional link between gene transcriptional state and SG biogenesis. Collectively these data demonstrate that ER-targeted and cytosolic mRNAs can be recruited into ER-associated SGs and this recruitment is sensitive to transcriptional inhibition. We propose that newly transcribed mRNAs exported under conditions of suppressed translation initiation are primary SG substrates, with the ER serving as the central subcellular site of SG formation.
Keywords: endoplasmic reticulum; mRNA; oxidative stress; stress granule; translational regulation; unfolded protein response.
© 2021 Child et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
Single-Molecule Imaging Reveals Translation of mRNAs Localized to Stress Granules.Cell. 2020 Dec 23;183(7):1801-1812.e13. doi: 10.1016/j.cell.2020.11.010. Epub 2020 Dec 11. Cell. 2020. PMID: 33308477
-
An emerging role for the endoplasmic reticulum in stress granule biogenesis.Semin Cell Dev Biol. 2024 Mar 15;156:160-166. doi: 10.1016/j.semcdb.2022.09.013. Epub 2022 Oct 4. Semin Cell Dev Biol. 2024. PMID: 36202692 Review.
-
Triptolide activates unfolded protein response leading to chronic ER stress in pancreatic cancer cells.Am J Physiol Gastrointest Liver Physiol. 2014 Jun 1;306(11):G1011-20. doi: 10.1152/ajpgi.00466.2013. Epub 2014 Apr 3. Am J Physiol Gastrointest Liver Physiol. 2014. PMID: 24699326 Free PMC article.
-
De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum.RNA. 2014 Oct;20(10):1489-98. doi: 10.1261/rna.045526.114. Epub 2014 Aug 20. RNA. 2014. PMID: 25142066 Free PMC article.
-
Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma.FEBS J. 2013 Sep;280(18):4348-70. doi: 10.1111/febs.12287. Epub 2013 May 9. FEBS J. 2013. PMID: 23587065 Review.
Cited by
-
The uncharted territory of host-pathogen interaction in tuberculosis.Front Immunol. 2024 Jan 19;15:1339467. doi: 10.3389/fimmu.2024.1339467. eCollection 2024. Front Immunol. 2024. PMID: 38312835 Free PMC article. Review.
-
Dynamics of nucleic acid mobility.Genetics. 2023 Aug 31;225(1):iyad132. doi: 10.1093/genetics/iyad132. Genetics. 2023. PMID: 37491977 Free PMC article.
-
Oligoadenylate synthetase 1 displays dual antiviral mechanisms in driving translational shutdown and protecting interferon production.Immunity. 2024 Mar 12;57(3):446-461.e7. doi: 10.1016/j.immuni.2024.02.002. Epub 2024 Feb 28. Immunity. 2024. PMID: 38423012
-
The enigma of ultraviolet radiation stress granules: Research challenges and new perspectives.Front Mol Biosci. 2022 Dec 1;9:1066650. doi: 10.3389/fmolb.2022.1066650. eCollection 2022. Front Mol Biosci. 2022. PMID: 36533077 Free PMC article.
-
Newly synthesized mRNA escapes translational repression during the acute phase of the mammalian unfolded protein response.PLoS One. 2022 Aug 10;17(8):e0271695. doi: 10.1371/journal.pone.0271695. eCollection 2022. PLoS One. 2022. PMID: 35947624 Free PMC article.
References
-
- Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Pietrement O, Pastre D. 2014. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res 42: 8678–8691. 10.1093/nar/gku582 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
