A split protease-E. coli ClpXP system quantifies protein-protein interactions in Escherichia coli cells
- PMID: 34230602
- PMCID: PMC8260793
- DOI: 10.1038/s42003-021-02374-w
A split protease-E. coli ClpXP system quantifies protein-protein interactions in Escherichia coli cells
Abstract
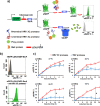
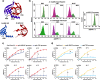
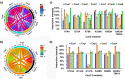
Characterizing protein-protein interactions (PPIs) is an effective method to help explore protein function. Here, through integrating a newly identified split human Rhinovirus 3 C (HRV 3 C) protease, super-folder GFP (sfGFP), and ClpXP-SsrA protein degradation machinery, we developed a fluorescence-assisted single-cell methodology (split protease-E. coli ClpXP (SPEC)) to explore protein-protein interactions for both eukaryotic and prokaryotic species in E. coli cells. We firstly identified a highly efficient split HRV 3 C protease with high re-assembly ability and then incorporated it into the SPEC method. The SPEC method could convert the cellular protein-protein interaction to quantitative fluorescence signals through a split HRV 3 C protease-mediated proteolytic reaction with high efficiency and broad temperature adaptability. Using SPEC method, we explored the interactions among effectors of representative type I-E and I-F CRISPR/Cas complexes, which combining with subsequent studies of Cas3 mutations conferred further understanding of the functions and structures of CRISPR/Cas complexes.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
ClpXP and ClpAP control the Escherichia coli division protein ZapC by proteolysis.Microbiology (Reading). 2016 Jun;162(6):909-920. doi: 10.1099/mic.0.000278. Epub 2016 Mar 15. Microbiology (Reading). 2016. PMID: 26978224 Free PMC article.
-
Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP.Mol Microbiol. 2008 Nov;70(4):1012-25. doi: 10.1111/j.1365-2958.2008.06467.x. Epub 2008 Sep 22. Mol Microbiol. 2008. PMID: 18811726 Free PMC article.
-
Protein knots provide mechano-resilience to an AAA+ protease-mediated proteolysis with profound ATP energy expenses.Biochim Biophys Acta Proteins Proteom. 2020 Feb;1868(2):140330. doi: 10.1016/j.bbapap.2019.140330. Epub 2019 Nov 20. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 31756432
-
Proteolysis: Adaptor, adaptor, catch me a catch.Curr Biol. 2004 Nov 9;14(21):R924-6. doi: 10.1016/j.cub.2004.10.015. Curr Biol. 2004. PMID: 15530384 Review.
-
Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli.Res Microbiol. 2009 Nov;160(9):667-76. doi: 10.1016/j.resmic.2009.08.014. Epub 2009 Sep 16. Res Microbiol. 2009. PMID: 19765651 Review.
Cited by
-
Genomic and epigenetic landscapes drive CRISPR-based genome editing in Bifidobacterium.Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2205068119. doi: 10.1073/pnas.2205068119. Epub 2022 Jul 20. Proc Natl Acad Sci U S A. 2022. PMID: 35857876 Free PMC article.
-
Direct and Ultrasensitive Bioluminescent Detection of Intact Respiratory Viruses.ACS Sens. 2024 Oct 25;9(10):5550-5560. doi: 10.1021/acssensors.4c01855. Epub 2024 Oct 7. ACS Sens. 2024. PMID: 39375866 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

