Gut microbiota signature of pathogen-dependent dysbiosis in viral gastroenteritis
- PMID: 34230563
- PMCID: PMC8260788
- DOI: 10.1038/s41598-021-93345-y
Gut microbiota signature of pathogen-dependent dysbiosis in viral gastroenteritis
Abstract
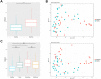
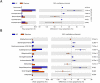
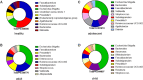
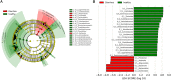
Acute gastroenteritis associated with diarrhea is considered a serious disease in Africa and South Asia. In this study, we examined the trends in the causative pathogens of diarrhea and the corresponding gut microbiota in Ghana using microbiome analysis performed on diarrheic stools via 16S rRNA sequencing. In total, 80 patients with diarrhea and 34 healthy adults as controls, from 2017 to 2018, were enrolled in the study. Among the patients with diarrhea, 39 were norovirus-positive and 18 were rotavirus-positive. The analysis of species richness (Chao1) was lower in patients with diarrhea than that in controls. Beta-diversity analysis revealed significant differences between the two groups. Several diarrhea-related pathogens (e.g., Escherichia-Shigella, Klebsiella and Campylobacter) were detected in patients with diarrhea. Furthermore, co-infection with these pathogens and enteroviruses (e.g., norovirus and rotavirus) was observed in several cases. Levels of both Erysipelotrichaceae and Staphylococcaceae family markedly differed between norovirus-positive and -negative diarrheic stools, and the 10 predicted metabolic pathways, including the carbohydrate metabolism pathway, showed significant differences between rotavirus-positive patients with diarrhea and controls. This comparative study of diarrheal pathogens in Ghana revealed specific trends in the gut microbiota signature associated with diarrhea and that pathogen-dependent dysbiosis occurred in viral gastroenteritis.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Mixed Viral-Bacterial Infections and Their Effects on Gut Microbiota and Clinical Illnesses in Children.Sci Rep. 2019 Jan 29;9(1):865. doi: 10.1038/s41598-018-37162-w. Sci Rep. 2019. PMID: 30696865 Free PMC article.
-
Investigation of gut microbiota diversity according to infectious agent in pediatric infectious acute gastroenteritis in a Korean university hospital.Pediatr Neonatol. 2024 Sep;65(5):476-481. doi: 10.1016/j.pedneo.2024.01.005. Epub 2024 Mar 6. Pediatr Neonatol. 2024. PMID: 38471992
-
Characterization of the human gut microbiome during travelers' diarrhea.Gut Microbes. 2015;6(2):110-9. doi: 10.1080/19490976.2015.1019693. Gut Microbes. 2015. PMID: 25695334 Free PMC article.
-
Rotavirus-mediated alteration of gut microbiota and its correlation with physiological characteristics in neonatal calves.J Microbiol. 2019 Feb;57(2):113-121. doi: 10.1007/s12275-019-8549-1. Epub 2018 Nov 19. J Microbiol. 2019. PMID: 30456757 Free PMC article.
-
Vaccines for viral and bacterial pathogens causing acute gastroenteritis: Part I: Overview, vaccines for enteric viruses and Vibrio cholerae.Hum Vaccin Immunother. 2015;11(3):584-600. doi: 10.1080/21645515.2015.1011019. Hum Vaccin Immunother. 2015. PMID: 25715048 Free PMC article. Review.
Cited by
-
Role of Microbiota in Viral Infections and Pathological Progression.Viruses. 2022 May 1;14(5):950. doi: 10.3390/v14050950. Viruses. 2022. PMID: 35632692 Free PMC article. Review.
-
Diarrhea-Causing Bacteria and Their Antibiotic Resistance Patterns Among Diarrhea Patients From Ghana.Front Microbiol. 2022 May 19;13:894319. doi: 10.3389/fmicb.2022.894319. eCollection 2022. Front Microbiol. 2022. PMID: 35663873 Free PMC article.
-
Minimal Impact on the Resistome of Children in Botswana After Azithromycin Treatment for Acute Severe Diarrheal Disease.J Infect Dis. 2024 Jul 25;230(1):239-249. doi: 10.1093/infdis/jiae049. J Infect Dis. 2024. PMID: 39052715 Clinical Trial.
-
Assessing Gut Microbiota in an Infant with Congenital Propionic Acidemia before and after Probiotic Supplementation.Microorganisms. 2021 Dec 16;9(12):2599. doi: 10.3390/microorganisms9122599. Microorganisms. 2021. PMID: 34946200 Free PMC article.
-
Animal and In Vitro Models as Powerful Tools to Decipher the Effects of Enteric Pathogens on the Human Gut Microbiota.Microorganisms. 2023 Dec 29;12(1):67. doi: 10.3390/microorganisms12010067. Microorganisms. 2023. PMID: 38257894 Free PMC article. Review.
References
-
- Ritchie, B. D. A. H. Diarrheal diseases. Published online at OurWorldInData.org. https://ourworldindata.org/diarrheal-diseases (2018).
-
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization-Coordinated Global Rotavirus Surveillance Network Global, regional, and national estimates of rotavirus mortality in children < 5 years of age, 2000–2013. Clin. Infect. Dis. 2016;62(2):S96–S105. doi: 10.1093/cid/civ1013. - DOI - PubMed

