HOME-BIO (sHOtgun MEtagenomic analysis of BIOlogical entities): a specific and comprehensive pipeline for metagenomic shotgun sequencing data analysis
- PMID: 34225648
- PMCID: PMC8256542
- DOI: 10.1186/s12859-021-04004-y
HOME-BIO (sHOtgun MEtagenomic analysis of BIOlogical entities): a specific and comprehensive pipeline for metagenomic shotgun sequencing data analysis
Abstract
Background: Next-Generation-Sequencing (NGS) enables detection of microorganisms present in biological and other matrices of various origin and nature, allowing not only the identification of known phyla and strains but also the discovery of novel ones. The large amount of metagenomic shotgun data produced by NGS require comprehensive and user-friendly pipelines for data analysis, that speed up the bioinformatics steps, relieving the users from the need to manually perform complex and time-consuming tasks.
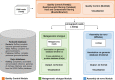
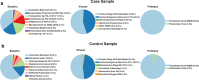
Results: We describe here HOME-BIO (sHOtgun MEtagenomic analysis of BIOlogical entities), an exhaustive pipeline for metagenomics data analysis, comprising three independent analytical modules designed for an inclusive analysis of large NGS datasets.
Conclusions: HOME-BIO is a powerful and easy-to-use tool that can be run also by users with limited computational expertise. It allows in-depth analyses by removing low-complexity/ problematic reads, integrating the analytical steps that lead to a comprehensive taxonomy profile of each sample by querying different source databases, and it is customizable according to specific users' needs.
Keywords: Next-Generation-Sequencing; Pipeline; Shotgun metagenomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments.Microbiome. 2019 Mar 22;7(1):46. doi: 10.1186/s40168-019-0658-x. Microbiome. 2019. PMID: 30902113 Free PMC article.
-
MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis.Microbiome. 2018 Sep 15;6(1):158. doi: 10.1186/s40168-018-0541-1. Microbiome. 2018. PMID: 30219103 Free PMC article.
-
From Whole Genome Shotgun Sequencing to Viral Community Profiling: The ViromeScan Tool.Methods Mol Biol. 2024;2732:23-28. doi: 10.1007/978-1-0716-3515-5_2. Methods Mol Biol. 2024. PMID: 38060115
-
[A review on the bioinformatics pipelines for metagenomic research].Dongwuxue Yanjiu. 2012 Dec;33(6):574-85. doi: 10.3724/SP.J.1141.2012.06574. Dongwuxue Yanjiu. 2012. PMID: 23266976 Review. Chinese.
-
Bioinformatics for NGS-based metagenomics and the application to biogas research.J Biotechnol. 2017 Nov 10;261:10-23. doi: 10.1016/j.jbiotec.2017.08.012. Epub 2017 Aug 18. J Biotechnol. 2017. PMID: 28823476 Review.
Cited by
-
Influence of Mycobiota in the Nasopharyngeal Tract of COVID-19 Patients.Microorganisms. 2024 Jul 19;12(7):1468. doi: 10.3390/microorganisms12071468. Microorganisms. 2024. PMID: 39065235 Free PMC article.
-
Tapping the Role of Microbial Biosurfactants in Pesticide Remediation: An Eco-Friendly Approach for Environmental Sustainability.Front Microbiol. 2021 Dec 23;12:791723. doi: 10.3389/fmicb.2021.791723. eCollection 2021. Front Microbiol. 2021. PMID: 35003022 Free PMC article. Review.
-
NGS analysis of nasopharyngeal microbiota in SARS-CoV-2 positive patients during the first year of the pandemic in the Campania Region of Italy.Microb Pathog. 2022 Apr;165:105506. doi: 10.1016/j.micpath.2022.105506. Epub 2022 Mar 28. Microb Pathog. 2022. PMID: 35358660 Free PMC article.
-
Dynamics of nasopharyngeal tract phageome and association with disease severity and age of patients during three waves of COVID-19.J Med Virol. 2022 Nov;94(11):5567-5573. doi: 10.1002/jmv.27998. Epub 2022 Jul 25. J Med Virol. 2022. PMID: 35831579 Free PMC article.
-
First Glimpse into the Genomic Characterization of People from the Imperial Roman Community of Casal Bertone (Rome, First-Third Centuries AD).Genes (Basel). 2022 Jan 13;13(1):136. doi: 10.3390/genes13010136. Genes (Basel). 2022. PMID: 35052476 Free PMC article.
References
-
- Sudarikov K, Tyakht A, Alexeev D. Methods for the metagenomic data visualization and analysis. Vol. 24, Current issues in molecular biology. Curr Issues Mol Biol; 2017. p. 37–58. - PubMed
-
- Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Vol. 35, Nature Biotechnology. Nature Publishing Group, Berlin; 2017. p. 833–44. - PubMed
-
- Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, et al. Culturing the human microbiota and culturomics. Vol. 16, Nature Reviews Microbiology. Nature Publishing Group, Berlin; 2018. p. 540–50. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

