Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis
- PMID: 34218797
- PMCID: PMC8256614
- DOI: 10.1186/s12872-021-02146-8
Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis
Abstract
Introduction: Heart failure (HF) is a heterogeneous clinical syndrome and affects millions of people all over the world. HF occurs when the cardiac overload and injury, which is a worldwide complaint. The aim of this study was to screen and verify hub genes involved in developmental HF as well as to explore active drug molecules.
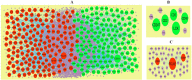
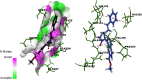
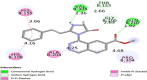
Methods: The expression profiling by high throughput sequencing of GSE141910 dataset was downloaded from the Gene Expression Omnibus (GEO) database, which contained 366 samples, including 200 heart failure samples and 166 non heart failure samples. The raw data was integrated to find differentially expressed genes (DEGs) and were further analyzed with bioinformatics analysis. Gene ontology (GO) and REACTOME enrichment analyses were performed via ToppGene; protein-protein interaction (PPI) networks of the DEGs was constructed based on data from the HiPPIE interactome database; modules analysis was performed; target gene-miRNA regulatory network and target gene-TF regulatory network were constructed and analyzed; hub genes were validated; molecular docking studies was performed.
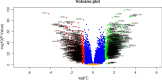
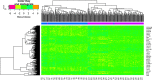
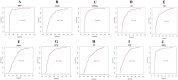
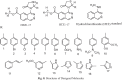
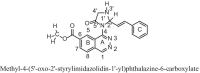
Results: A total of 881 DEGs, including 442 up regulated genes and 439 down regulated genes were observed. Most of the DEGs were significantly enriched in biological adhesion, extracellular matrix, signaling receptor binding, secretion, intrinsic component of plasma membrane, signaling receptor activity, extracellular matrix organization and neutrophil degranulation. The top hub genes ESR1, PYHIN1, PPP2R2B, LCK, TP63, PCLAF, CFTR, TK1, ECT2 and FKBP5 were identified from the PPI network. Module analysis revealed that HF was associated with adaptive immune system and neutrophil degranulation. The target genes, miRNAs and TFs were identified from the target gene-miRNA regulatory network and target gene-TF regulatory network. Furthermore, receiver operating characteristic (ROC) curve analysis and RT-PCR analysis revealed that ESR1, PYHIN1, PPP2R2B, LCK, TP63, PCLAF, CFTR, TK1, ECT2 and FKBP5 might serve as prognostic, diagnostic biomarkers and therapeutic target for HF. The predicted targets of these active molecules were then confirmed.
Conclusion: The current investigation identified a series of key genes and pathways that might be involved in the progression of HF, providing a new understanding of the underlying molecular mechanisms of HF.
Keywords: Differentially expressed genes; Enrichment analysis; Heart failure; Molecular docking; Prognosis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Identification of key pathways and genes in polycystic ovary syndrome via integrated bioinformatics analysis and prediction of small therapeutic molecules.Reprod Biol Endocrinol. 2021 Feb 23;19(1):31. doi: 10.1186/s12958-021-00706-3. Reprod Biol Endocrinol. 2021. PMID: 33622336 Free PMC article.
-
Identification of common signature genes and pathways underlying the pathogenesis association between nonalcoholic fatty liver disease and heart failure.Front Immunol. 2024 Sep 16;15:1424308. doi: 10.3389/fimmu.2024.1424308. eCollection 2024. Front Immunol. 2024. PMID: 39351239 Free PMC article.
-
Identification of biomarkers, pathways, and potential therapeutic targets for heart failure using next-generation sequencing data and bioinformatics analysis.Ther Adv Cardiovasc Dis. 2023 Jan-Dec;17:17539447231168471. doi: 10.1177/17539447231168471. Ther Adv Cardiovasc Dis. 2023. PMID: 37092838 Free PMC article.
-
STAT4 and COL1A2 are potential diagnostic biomarkers and therapeutic targets for heart failure comorbided with depression.Brain Res Bull. 2022 Jun 15;184:68-75. doi: 10.1016/j.brainresbull.2022.03.014. Epub 2022 Mar 31. Brain Res Bull. 2022. PMID: 35367598 Review.
-
Potential Mechanisms Between HF and COPD: New Insights From Bioinformatics.Curr Probl Cardiol. 2023 Mar;48(3):101539. doi: 10.1016/j.cpcardiol.2022.101539. Epub 2022 Dec 15. Curr Probl Cardiol. 2023. PMID: 36528207 Review.
Cited by
-
Identification of novel biomarkers involved in doxorubicin-induced acute and chronic cardiotoxicity, respectively, by integrated bioinformatics.Front Cardiovasc Med. 2023 Jan 11;9:996809. doi: 10.3389/fcvm.2022.996809. eCollection 2022. Front Cardiovasc Med. 2023. PMID: 36712272 Free PMC article.
-
Alterated gene expression in dilated cardiomyopathy after left ventricular assist device support by bioinformatics analysis.Front Cardiovasc Med. 2023 Mar 17;10:1013057. doi: 10.3389/fcvm.2023.1013057. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37008310 Free PMC article.
-
Machine Learning in Identifying Marker Genes for Congenital Heart Diseases of Different Cardiac Cell Types.Life (Basel). 2024 Aug 19;14(8):1032. doi: 10.3390/life14081032. Life (Basel). 2024. PMID: 39202774 Free PMC article.
-
Connections for Matters of the Heart: Network Medicine in Cardiovascular Diseases.Front Cardiovasc Med. 2022 May 19;9:873582. doi: 10.3389/fcvm.2022.873582. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35665246 Free PMC article. Review.
-
Myocardial RNA Sequencing Reveals New Potential Therapeutic Targets in Heart Failure with Preserved Ejection Fraction.Biomedicines. 2023 Jul 28;11(8):2131. doi: 10.3390/biomedicines11082131. Biomedicines. 2023. PMID: 37626628 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

