LAMC1 upregulation via TGFβ induces inflammatory cancer-associated fibroblasts in esophageal squamous cell carcinoma via NF-κB-CXCL1-STAT3
- PMID: 34218518
- PMCID: PMC8564640
- DOI: 10.1002/1878-0261.13053
LAMC1 upregulation via TGFβ induces inflammatory cancer-associated fibroblasts in esophageal squamous cell carcinoma via NF-κB-CXCL1-STAT3
Abstract
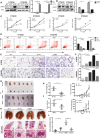
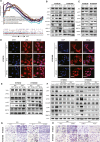
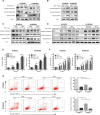
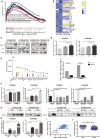
Cancer-associated fibroblasts (CAF) are a heterogeneous cell population within the tumor microenvironment,and play an important role in tumor development. By regulating the heterogeneity of CAF, transforming growth factor β (TGFβ) influences tumor development. Here, we explored oncogenes regulated by TGFβ1 that are also involved in signaling pathways and interactions within the tumor microenvironment. We analyzed sequencing data of The Cancer Genome Atlas (TCGA) and our own previously established RNA microarray data (GSE53625), as well as esophageal squamous cell carcinoma (ESCC) cell lines with or without TGFβ1 stimulation. We then focused on laminin subunit gamma 1 (LAMC1), which was overexpressed in ESCC cells, affecting patient prognosis, which could be upregulated by TGFβ1 through the synergistic activation of SMAD family member 4 (SMAD4) and SP1. LAMC1 directly promoted the proliferation and migration of tumor cells, mainly via Akt-NFκB-MMP9/14 signaling. Additionally, LAMC1 promoted CXCL1 secretion, which stimulated the formation of inflammatory CAF (iCAF) through CXCR2-pSTAT3. Inflammatory CAF promoted tumor progression. In summary, we identified the dual mechanism by which the upregulation of LAMC1 by TGFβ in tumor cells not only promotes ESCC proliferation and migration, but also indirectly induces carcinogenesis by stimulating CXCL1 secretion to promote the formation of iCAF. This finding suggests that LAMC1 could be a potential therapeutic target and prognostic marker for ESCC.
Keywords: CAF; CXCL1; ESCC; LAMC1; heterogeneity; transforming growth factor β.
© 2021 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
AREG Upregulation in Cancer Cells via Direct Interaction with Cancer-Associated Fibroblasts Promotes Esophageal Squamous Cell Carcinoma Progression Through EGFR-Erk/p38 MAPK Signaling.Cells. 2024 Oct 19;13(20):1733. doi: 10.3390/cells13201733. Cells. 2024. PMID: 39451251 Free PMC article.
-
Collagen 1-mediated CXCL1 secretion in tumor cells activates fibroblasts to promote radioresistance of esophageal cancer.Cell Rep. 2023 Oct 31;42(10):113270. doi: 10.1016/j.celrep.2023.113270. Epub 2023 Oct 17. Cell Rep. 2023. PMID: 37851572
-
CAF-secreted CXCL1 conferred radioresistance by regulating DNA damage response in a ROS-dependent manner in esophageal squamous cell carcinoma.Cell Death Dis. 2017 May 18;8(5):e2790. doi: 10.1038/cddis.2017.180. Cell Death Dis. 2017. PMID: 28518141 Free PMC article.
-
Emerging role of cancer-associated fibroblasts in esophageal squamous cell carcinoma.Pathol Res Pract. 2024 Jan;253:155002. doi: 10.1016/j.prp.2023.155002. Epub 2023 Nov 30. Pathol Res Pract. 2024. PMID: 38056131 Review.
-
Cancer-associated fibroblasts: An emerging target against esophageal squamous cell carcinoma.Cancer Lett. 2022 Oct 10;546:215860. doi: 10.1016/j.canlet.2022.215860. Epub 2022 Aug 7. Cancer Lett. 2022. PMID: 35948121 Review.
Cited by
-
CAFs-derived Exosomal miR-889-3p Might Repress M1 Macrophage Polarization to Boost ESCC Development by Regulating STAT1.Cell Biochem Biophys. 2024 Sep 5. doi: 10.1007/s12013-024-01496-2. Online ahead of print. Cell Biochem Biophys. 2024. PMID: 39237779
-
Identification of fibroblast-related genes based on single-cell and machine learning to predict the prognosis and endocrine metabolism of pancreatic cancer.Front Endocrinol (Lausanne). 2023 Jul 31;14:1201755. doi: 10.3389/fendo.2023.1201755. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37588985 Free PMC article.
-
The axis of tumor-associated macrophages, extracellular matrix proteins, and cancer-associated fibroblasts in oncogenesis.Cancer Cell Int. 2024 Oct 7;24(1):335. doi: 10.1186/s12935-024-03518-8. Cancer Cell Int. 2024. PMID: 39375726 Free PMC article. Review.
-
Cancer-Associated Fibroblasts Hinder Lung Squamous Cell Carcinoma Oxidative Stress-Induced Apoptosis via METTL3 Mediated m6A Methylation of COL10A1.Oxid Med Cell Longev. 2022 Oct 6;2022:4320809. doi: 10.1155/2022/4320809. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36246404 Free PMC article.
-
Identification of basement membrane markers in diabetic kidney disease and immune infiltration by using bioinformatics analysis and experimental verification.IET Syst Biol. 2023 Dec;17(6):316-326. doi: 10.1049/syb2.12078. Epub 2023 Sep 30. IET Syst Biol. 2023. PMID: 37776100 Free PMC article.
References
-
- Lagergren J, Smyth E, Cunningham D & Lagergren P (2017) Oesophageal cancer. Lancet 390, 2383–2396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

