De novo transcriptome analysis of white teak (Gmelina arborea Roxb) wood reveals critical genes involved in xylem development and secondary metabolism
- PMID: 34215181
- PMCID: PMC8252223
- DOI: 10.1186/s12864-021-07777-x
De novo transcriptome analysis of white teak (Gmelina arborea Roxb) wood reveals critical genes involved in xylem development and secondary metabolism
Abstract
Background: Gmelina arborea Roxb is a fast-growing tree species of commercial importance for tropical countries due to multiple industrial uses of its wood. Wood is primarily composed of thick secondary cell walls of xylem cells which imparts the strength to the wood. Identification of the genes involved in the secondary cell wall biosynthesis as well as their cognate regulators is crucial to understand how the production of wood occurs and serves as a starting point for developing breeding strategies to produce varieties with improved wood quality, better paper pulping or new potential uses such as biofuel production. In order to gain knowledge on the molecular mechanisms and gene regulation related with wood development in white teak, a de novo sequencing and transcriptome assembly approach was used employing secondary cell wall synthesizing cells from young white teak trees.
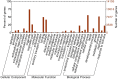
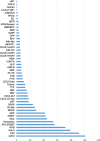
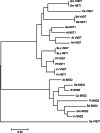
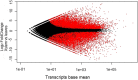
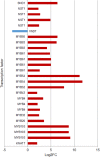
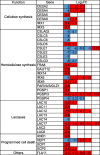
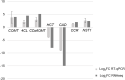
Results: For generation of transcriptome, RNA-seq reads were assembled into 110,992 transcripts and 49,364 genes were functionally annotated using plant databases; 5071 GO terms and 25,460 SSR markers were identified within xylem transcripts and 10,256 unigenes were assigned to KEGG database in 130 pathways. Among transcription factor families, C2H2, C3H, bLHLH and MYB were the most represented in xylem. Differential gene expression analysis using leaves as a reference was carried out and a total of 20,954 differentially expressed genes were identified including monolignol biosynthetic pathway genes. The differential expression of selected genes (4CL, COMT, CCoAOMT, CCR and NST1) was validated using qPCR.
Conclusions: We report the very first de novo transcriptome of xylem-related genes in this tropical timber species of commercial importance and constitutes a valuable extension of the publicly available transcriptomic resource aimed at fostering both basic and breeding studies.
Keywords: Differential gene expression; RNA-seq; Wood development; Xylem.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Large-scale transcriptional profiling of lignified tissues in Tectona grandis.BMC Plant Biol. 2015 Sep 15;15:221. doi: 10.1186/s12870-015-0599-x. BMC Plant Biol. 2015. PMID: 26369560 Free PMC article.
-
Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don.BMC Genomics. 2009 Jan 21;10:41. doi: 10.1186/1471-2164-10-41. BMC Genomics. 2009. PMID: 19159482 Free PMC article.
-
Wood transcriptome analysis and expression variation of lignin biosynthetic pathway transcripts in Ailanthus excelsa Roxb., a multi-purpose tropical tree species.J Biosci. 2021;46:105. J Biosci. 2021. PMID: 34840147
-
Wood of trees: Cellular structure, molecular formation, and genetic engineering.J Integr Plant Biol. 2024 Mar;66(3):443-467. doi: 10.1111/jipb.13589. Epub 2024 Jan 9. J Integr Plant Biol. 2024. PMID: 38032010 Review.
-
The transcriptomics of secondary growth and wood formation in conifers.Mol Biol Int. 2013;2013:974324. doi: 10.1155/2013/974324. Epub 2013 Oct 29. Mol Biol Int. 2013. PMID: 24288610 Free PMC article. Review.
Cited by
-
De novo transcriptome sequencing of Impatiens uliginosa and the analysis of candidate genes related to spur development.BMC Plant Biol. 2022 Dec 1;22(1):553. doi: 10.1186/s12870-022-03894-1. BMC Plant Biol. 2022. PMID: 36456926 Free PMC article.
References
-
- Hinchee M, Rottmann W, Mullinax L, Zhang C, Chang S, Cunningham M, et al. Short-rotation woody crops for bioenergy and biofuels applications. In Vitro Cell Dev Biol Plant. 2009;45(6):619–29. Epub 2009/11/26. 10.1007/s11627-009-9235-5. PubMed PMID: 19936031; PubMed Central PMCID: PMCPMC2778772. - PMC - PubMed
-
- Zhang G, Wang L, Li X, Bai S, Li Z, Yanting ST, et al. Distinctively altered lignin biosynthesis by site-modification of OsCAD2 for enhanced biomass saccharification in rice. GCB Bioenergy. 2020;13:305–319. doi: 10.1111/gcbb.12772. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

