AP-2α-Mediated Activation of E2F and EZH2 Drives Melanoma Metastasis
- PMID: 34210752
- PMCID: PMC8416798
- DOI: 10.1158/0008-5472.CAN-21-0772
AP-2α-Mediated Activation of E2F and EZH2 Drives Melanoma Metastasis
Abstract
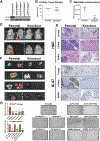
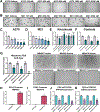
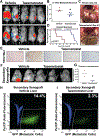
In melanoma metastasis, the role of the AP-2α transcription factor, which is encoded by TFAP2A, is controversial as some findings have suggested tumor suppressor activity while other studies have shown high TFAP2A expression in node-positive melanoma associated with poor prognosis. Here we demonstrate that AP-2α facilitates melanoma metastasis through transcriptional activation of genes within the E2F pathway including EZH2. A BioID screen found that AP-2α interacts with members of the nucleosome remodeling and deacetylase (NuRD) complex. Loss of AP-2α removed activating chromatin marks in the promoters of EZH2 and other E2F target genes through activation of the NuRD repression complex. In melanoma cells, treatment with tazemetostat, an FDA-approved and highly specific EZH2 inhibitor, substantially reduced anchorage-independent colony formation and demonstrated heritable antimetastatic effects, which were dependent on AP-2α. Single-cell RNA sequencing analysis of a metastatic melanoma mouse model revealed hyperexpansion of Tfap2a High/E2F-activated cell populations in transformed melanoma relative to progenitor melanocyte stem cells. These findings demonstrate that melanoma metastasis is driven by the AP-2α/EZH2 pathway and suggest that AP-2α expression can be used as a biomarker to predict responsiveness to EZH2 inhibitors for the treatment of advanced melanomas. SIGNIFICANCE: AP-2α drives melanoma metastasis by upregulating E2F pathway genes including EZH2 through inhibition of the NuRD repression complex, serving as a biomarker to predict responsiveness to EZH2 inhibitors.
©2021 American Association for Cancer Research.
Conflict of interest statement
All authors declare there are no competing financial interests in relation to the work described.
Figures




Similar articles
-
The EZH2 inhibitor tazemetostat upregulates the expression of CCL17/TARC in B-cell lymphoma and enhances T-cell recruitment.Cancer Sci. 2021 Nov;112(11):4604-4616. doi: 10.1111/cas.15122. Epub 2021 Sep 9. Cancer Sci. 2021. PMID: 34449935 Free PMC article.
-
Evaluation of an EZH2 inhibitor in patient-derived orthotopic xenograft models of pediatric brain tumors alone and in combination with chemo- and radiation therapies.Lab Invest. 2022 Feb;102(2):185-193. doi: 10.1038/s41374-021-00700-8. Epub 2021 Nov 20. Lab Invest. 2022. PMID: 34802040 Free PMC article.
-
Chromatin remodeling by the histone methyltransferase EZH2 drives lung pre-malignancy and is a target for cancer prevention.Clin Epigenetics. 2021 Feb 25;13(1):44. doi: 10.1186/s13148-021-01034-4. Clin Epigenetics. 2021. PMID: 33632299 Free PMC article.
-
Targeting PRC2 for the treatment of cancer: an updated patent review (2016 - 2020).Expert Opin Ther Pat. 2021 Feb;31(2):119-135. doi: 10.1080/13543776.2021.1841167. Epub 2021 Jan 4. Expert Opin Ther Pat. 2021. PMID: 33103538 Review.
-
Pharmacology and pharmacokinetics of tazemetostat.Cancer Chemother Pharmacol. 2024 May;93(5):509-517. doi: 10.1007/s00280-024-04658-4. Epub 2024 Mar 23. Cancer Chemother Pharmacol. 2024. PMID: 38520556 Review.
Cited by
-
Potential prognosis and immunotherapy predictor TFAP2A in pan-cancer.Aging (Albany NY). 2024 Jan 23;16(2):1021-1048. doi: 10.18632/aging.205225. Epub 2024 Jan 23. Aging (Albany NY). 2024. PMID: 38265973 Free PMC article.
-
TFAP2 paralogs facilitate chromatin access for MITF at pigmentation and cell proliferation genes.PLoS Genet. 2022 May 17;18(5):e1010207. doi: 10.1371/journal.pgen.1010207. eCollection 2022 May. PLoS Genet. 2022. PMID: 35580127 Free PMC article.
-
Pathological and Molecular Diagnosis of Uveal Melanoma.Diagnostics (Basel). 2024 May 2;14(9):958. doi: 10.3390/diagnostics14090958. Diagnostics (Basel). 2024. PMID: 38732371 Free PMC article. Review.
-
Construction and validation of a chromatin regulator-related gene signature for prognostic and therapeutic significance of clear cell renal cell carcinoma.Transl Cancer Res. 2024 Jan 31;13(1):150-172. doi: 10.21037/tcr-23-1383. Epub 2024 Jan 15. Transl Cancer Res. 2024. PMID: 38410230 Free PMC article.
-
Identification of genetic fingerprint of type I interferon therapy in visceral metastases of melanoma.Sci Rep. 2024 Nov 3;14(1):26540. doi: 10.1038/s41598-024-77285-x. Sci Rep. 2024. PMID: 39489756 Free PMC article.
References
-
- Spagnolo F, Boutros A, Tanda E, Queirolo P. The adjuvant treatment revolution for high-risk melanoma patients. Semin Cancer Biol 2019;59:283–9 - PubMed
-
- Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development 2004;131:1463–77 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

