The Central Role of Cytochrome P450 in Xenobiotic Metabolism-A Brief Review on a Fascinating Enzyme Family
- PMID: 34206277
- PMCID: PMC8293344
- DOI: 10.3390/jox11030007
The Central Role of Cytochrome P450 in Xenobiotic Metabolism-A Brief Review on a Fascinating Enzyme Family
Abstract
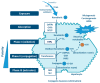
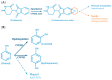
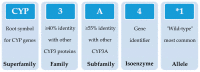
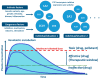
Human Cytochrome P450 (CYP) enzymes constitute a superfamily of membrane-bound hemoproteins that are responsible for the metabolism of a wide variety of clinically, physiologically, and toxicologically important compounds. These heme-thiolate monooxygenases play a pivotal role in the detoxification of xenobiotics, participating in the metabolism of many structurally diverge compounds. This short-review is intended to provide a summary on the major roles of CYPs in Phase I xenobiotic metabolism. The manuscript is focused on eight main topics that include the most relevant aspects of past and current CYP research. Initially, (I) a general overview of the main aspects of absorption, distribution, metabolism, and excretion (ADME) of xenobiotics are presented. This is followed by (II) a background overview on major achievements in the past of the CYP research field. (III) Classification and nomenclature of CYPs is briefly reviewed, followed by (IV) a summary description on CYP's location and function in mammals. Subsequently, (V) the physiological relevance of CYP as the cornerstone of Phase I xenobiotic metabolism is highlighted, followed by (VI) reviewing both genetic determinants and (VI) nongenetic factors in CYP function and activity. The last topic of the review (VIII) is focused on the current challenges of the CYP research field.
Keywords: Cytochrome P450 (CYP); adverse drug reactions (ADRs); carcinogens; drug-metabolizing enzymes (DMEs); metabolism; toxicology; xenobiotic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Pleiotropy of Progesterone Receptor Membrane Component 1 in Modulation of Cytochrome P450 Activity.J Xenobiot. 2024 May 1;14(2):575-603. doi: 10.3390/jox14020034. J Xenobiot. 2024. PMID: 38804287 Free PMC article. Review.
-
Cytochrome P450 enzymes and metabolism of drugs and neurotoxins within the mammalian brain.Adv Pharmacol. 2022;95:73-106. doi: 10.1016/bs.apha.2022.04.003. Epub 2022 Jun 10. Adv Pharmacol. 2022. PMID: 35953164
-
Cytochrome P450 Structure, Function and Clinical Significance: A Review.Curr Drug Targets. 2018;19(1):38-54. doi: 10.2174/1389450118666170125144557. Curr Drug Targets. 2018. PMID: 28124606 Review.
-
Update on allele nomenclature for human cytochromes P450 and the Human Cytochrome P450 Allele (CYP-allele) Nomenclature Database.Methods Mol Biol. 2013;987:251-9. doi: 10.1007/978-1-62703-321-3_21. Methods Mol Biol. 2013. PMID: 23475683
-
Xenobiotic-metabolizing cytochrome P450 enzymes in the human feto-placental unit: role in intrauterine toxicity.Crit Rev Toxicol. 1998 Jan;28(1):35-72. doi: 10.1080/10408449891344173. Crit Rev Toxicol. 1998. PMID: 9493761 Review.
Cited by
-
Synthesis of mono Cytochrome P450 in a modified CHO-CPR cell-free protein production platform.Sci Rep. 2024 Jan 13;14(1):1271. doi: 10.1038/s41598-024-51781-6. Sci Rep. 2024. PMID: 38218994 Free PMC article.
-
Bisphenol A is a carcinogen that induces lipid accumulation, peroxisome proliferator‑activated receptor‑γ expression and liver disease.Exp Ther Med. 2022 Oct 31;24(6):735. doi: 10.3892/etm.2022.11671. eCollection 2022 Dec. Exp Ther Med. 2022. PMID: 36466761 Free PMC article.
-
Pharmacogenetic Variation in Neanderthals and Denisovans and Implications for Human Health and Response to Medications.Genome Biol Evol. 2023 Dec 1;15(12):evad222. doi: 10.1093/gbe/evad222. Genome Biol Evol. 2023. PMID: 38051947 Free PMC article.
-
Cytochromes P450 involved in bacterial RiPP biosyntheses.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuad005. doi: 10.1093/jimb/kuad005. J Ind Microbiol Biotechnol. 2023. PMID: 36931895 Free PMC article. Review.
-
The Role of Cytochrome P450 Enzymes in COVID-19 Pathogenesis and Therapy.Front Pharmacol. 2022 Feb 2;13:791922. doi: 10.3389/fphar.2022.791922. eCollection 2022. Front Pharmacol. 2022. PMID: 35185562 Free PMC article. Review.
References
-
- Croom E. Metabolism of Xenobiotics of Human Environments. In: Ernest H., editor. Progress in Molecular Biology and Translational Science. Volume 112. Elsevier; Amsterdam, The Netherlands: 2012. pp. 31–88. - PubMed
-
- Juchau M.R., Chen H. Developmental Enzymology. In: William S. Jr., Louis W.C., editors. Handbook of Developmental Neurotoxicology. Elsevier; Amsterdam, The Netherlands: 1998. pp. 321–337.
-
- Evans T.J. Chapter 2—Toxicokinetics and Toxicodynamics. In: Peterson M.E., Talcott P.A., editors. Small Animal Toxicology. 3rd ed. W.B. Saunders; Saint Louis, MO, USA: 2013. pp. 13–19.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

