The Functional Consequences of the Novel Ribosomal Pausing Site in SARS-CoV-2 Spike Glycoprotein RNA
- PMID: 34204305
- PMCID: PMC8235447
- DOI: 10.3390/ijms22126490
The Functional Consequences of the Novel Ribosomal Pausing Site in SARS-CoV-2 Spike Glycoprotein RNA
Abstract
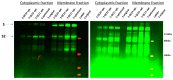
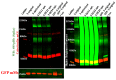
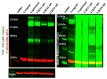
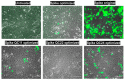
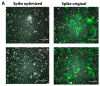
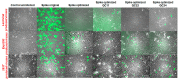
The SARS-CoV-2 Spike glycoprotein (S protein) acquired a unique new 4 amino acid -PRRA- insertion sequence at amino acid residues (aa) 681-684 that forms a new furin cleavage site in S protein as well as several new glycosylation sites. We studied various statistical properties of the -PRRA- insertion at the RNA level (CCUCGGCGGGCA). The nucleotide composition and codon usage of this sequence are different from the rest of the SARS-CoV-2 genome. One of such features is two tandem CGG codons, although the CGG codon is the rarest codon in the SARS-CoV-2 genome. This suggests that the insertion sequence could cause ribosome pausing as the result of these rare codons. Due to population variants, the Nextstrain divergence measure of the CCU codon is extremely large. We cannot exclude that this divergence might affect host immune responses/effectiveness of SARS-CoV-2 vaccines, possibilities awaiting further investigation. Our experimental studies show that the expression level of original RNA sequence "wildtype" spike protein is much lower than for codon-optimized spike protein in all studied cell lines. Interestingly, the original spike sequence produces a higher titer of pseudoviral particles and a higher level of infection. Further mutagenesis experiments suggest that this dual-effect insert, comprised of a combination of overlapping translation pausing and furin sites, has allowed SARS-CoV-2 to infect its new host (human) more readily. This underlines the importance of ribosome pausing to allow efficient regulation of protein expression and also of cotranslational subdomain folding.
Keywords: SARS-CoV-2; codon usage; ribosome pausing site; ribosome stalling; spike protein.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




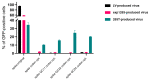
Similar articles
-
Probable human origin of the SARS-CoV-2 polybasic furin cleavage motif.BMC Genom Data. 2023 Nov 21;24(1):71. doi: 10.1186/s12863-023-01169-8. BMC Genom Data. 2023. PMID: 37990144 Free PMC article.
-
D614G Substitution of SARS-CoV-2 Spike Protein Increases Syncytium Formation and Virus Titer via Enhanced Furin-Mediated Spike Cleavage.mBio. 2021 Aug 31;12(4):e0058721. doi: 10.1128/mBio.00587-21. Epub 2021 Jul 27. mBio. 2021. PMID: 34311586 Free PMC article.
-
Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity.Nat Commun. 2024 May 16;15(1):4162. doi: 10.1038/s41467-024-48503-x. Nat Commun. 2024. PMID: 38755139 Free PMC article.
-
Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination.J Med Virol. 2022 May;94(5):1815-1820. doi: 10.1002/jmv.27539. Epub 2021 Dec 31. J Med Virol. 2022. PMID: 34936124 Review.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
Cited by
-
Many Faces of Next-Generation Sequencing in Gene Expression Studies.Int J Mol Sci. 2023 Feb 17;24(4):4075. doi: 10.3390/ijms24044075. Int J Mol Sci. 2023. PMID: 36835483 Free PMC article.
-
SARS-CoV-2 and Emerging Variants: Unmasking Structure, Function, Infection, and Immune Escape Mechanisms.Front Cell Infect Microbiol. 2022 May 12;12:869832. doi: 10.3389/fcimb.2022.869832. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646741 Free PMC article. Review.
-
Low-Density Lipoprotein Receptor (LDLR) Is Involved in Internalization of Lentiviral Particles Pseudotyped with SARS-CoV-2 Spike Protein in Ocular Cells.Int J Mol Sci. 2023 Jul 24;24(14):11860. doi: 10.3390/ijms241411860. Int J Mol Sci. 2023. PMID: 37511618 Free PMC article.
-
The Development of SARS-CoV-2 Variants: The Gene Makes the Disease.J Dev Biol. 2021 Dec 15;9(4):58. doi: 10.3390/jdb9040058. J Dev Biol. 2021. PMID: 34940505 Free PMC article. Review.
-
The 29-nucleotide deletion in SARS-CoV: truncated versions of ORF8 are under purifying selection.BMC Genomics. 2023 Jul 10;24(1):387. doi: 10.1186/s12864-023-09482-3. BMC Genomics. 2023. PMID: 37430204 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

