Immunogenomics of Colorectal Cancer Response to Checkpoint Blockade: Analysis of the KEYNOTE 177 Trial and Validation Cohorts
- PMID: 34197832
- PMCID: PMC8527923
- DOI: 10.1053/j.gastro.2021.06.064
Immunogenomics of Colorectal Cancer Response to Checkpoint Blockade: Analysis of the KEYNOTE 177 Trial and Validation Cohorts
Abstract
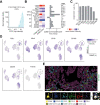
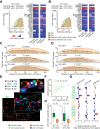
Background & aims: Colorectal cancer (CRC) shows variable response to immune checkpoint blockade, which can only partially be explained by high tumor mutational burden (TMB). We conducted an integrated study of the cancer tissue and associated tumor microenvironment (TME) from patients treated with pembrolizumab (KEYNOTE 177 clinical trial) or nivolumab to dissect the cellular and molecular determinants of response to anti- programmed cell death 1 (PD1) immunotherapy.
Methods: We selected multiple regions per tumor showing variable T-cell infiltration for a total of 738 regions from 29 patients, divided into discovery and validation cohorts. We performed multiregional whole-exome and RNA sequencing of the tumor cells and integrated these with T-cell receptor sequencing, high-dimensional imaging mass cytometry, detection of programmed death-ligand 1 (PDL1) interaction in situ, multiplexed immunofluorescence, and computational spatial analysis of the TME.
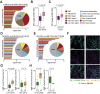
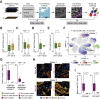
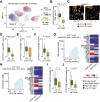
Results: In hypermutated CRCs, response to anti-PD1 immunotherapy was not associated with TMB but with high clonality of immunogenic mutations, clonally expanded T cells, low activation of Wnt signaling, deregulation of the interferon gamma pathway, and active immune escape mechanisms. Responsive hypermutated CRCs were also rich in cytotoxic and proliferating PD1+CD8 T cells interacting with PDL1+ antigen-presenting macrophages.
Conclusions: Our study clarified the limits of TMB as a predictor of response of CRC to anti-PD1 immunotherapy. It identified a population of antigen-presenting macrophages interacting with CD8 T cells that consistently segregate with response. We therefore concluded that anti-PD1 agents release the PD1-PDL1 interaction between CD8 T cells and macrophages to promote cytotoxic antitumor activity.
Keywords: Anti-PD1 Immunotherapy; CD8 T cells; Interferon Gamma; Tumor Mutational Burden; Wnt Signaling.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Dickkopf 1 impairs the tumor response to PD-1 blockade by inactivating CD8+ T cells in deficient mismatch repair colorectal cancer.J Immunother Cancer. 2021 Mar;9(3):e001498. doi: 10.1136/jitc-2020-001498. J Immunother Cancer. 2021. PMID: 33782107 Free PMC article.
-
Molecular correlates of response to nivolumab at baseline and on treatment in patients with RCC.J Immunother Cancer. 2021 Mar;9(3):e001506. doi: 10.1136/jitc-2020-001506. J Immunother Cancer. 2021. PMID: 33658305 Free PMC article. Clinical Trial.
-
A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study.Lancet Oncol. 2018 Sep;19(9):1180-1191. doi: 10.1016/S1470-2045(18)30413-3. Epub 2018 Aug 14. Lancet Oncol. 2018. PMID: 30120041
-
Cancer immunotherapy resistance based on immune checkpoints inhibitors: Targets, biomarkers, and remedies.Drug Resist Updat. 2020 Dec;53:100718. doi: 10.1016/j.drup.2020.100718. Epub 2020 Jul 15. Drug Resist Updat. 2020. PMID: 32736034 Review.
-
The Potential Value of Immunotherapy in Colorectal Cancers: Review of the Evidence for Programmed Death-1 Inhibitor Therapy.Clin Colorectal Cancer. 2016 Dec;15(4):285-291. doi: 10.1016/j.clcc.2016.07.007. Epub 2016 Jul 22. Clin Colorectal Cancer. 2016. PMID: 27553906 Review.
Cited by
-
The current landscape of spatial biomarkers for prediction of response to immune checkpoint inhibition.NPJ Precis Oncol. 2024 Aug 13;8(1):178. doi: 10.1038/s41698-024-00671-1. NPJ Precis Oncol. 2024. PMID: 39138341 Free PMC article. Review.
-
Identification of four immune subtypes in locally advanced rectal cancer treated with neoadjuvant chemotherapy for predicting the efficacy of subsequent immune checkpoint blockade.Front Immunol. 2022 Sep 27;13:955187. doi: 10.3389/fimmu.2022.955187. eCollection 2022. Front Immunol. 2022. PMID: 36238279 Free PMC article.
-
Comprehensive Bioinformatics Analysis of Gasdermin Family of Glioma.Comput Intell Neurosci. 2022 Apr 15;2022:9046507. doi: 10.1155/2022/9046507. eCollection 2022. Comput Intell Neurosci. 2022. Retraction in: Comput Intell Neurosci. 2023 Dec 13;2023:9852045. doi: 10.1155/2023/9852045 PMID: 35463276 Free PMC article. Retracted.
-
Potent therapeutic strategy in gastric cancer with microsatellite instability-high and/or deficient mismatch repair.Gastric Cancer. 2024 Sep;27(5):907-931. doi: 10.1007/s10120-024-01523-4. Epub 2024 Jun 26. Gastric Cancer. 2024. PMID: 38922524 Free PMC article. Review.
-
Cancer-associated foam cells hamper protective T cell immunity and favor tumor progression in human colon carcinogenesis.J Immunother Cancer. 2024 Oct 12;12(10):e009720. doi: 10.1136/jitc-2024-009720. J Immunother Cancer. 2024. PMID: 39395839 Free PMC article.
References
-
- Van den Eynde M., Mlecnik B., Bindea G. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell. 2018;34:1012–1026.e3. - PubMed
-
- André T., Shiu K.-K., Kim T.W. Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N Engl J Med. 2020;383:2207–2218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- C43634/A25487/CRUK_/Cancer Research UK/United Kingdom
- FC001745/ARC_/Arthritis Research UK/United Kingdom
- C604/A25135/CRUK_/Cancer Research UK/United Kingdom
- FC001169/ARC_/Arthritis Research UK/United Kingdom
- 25253/CRUK_/Cancer Research UK/United Kingdom
- 30025/CRUK_/Cancer Research UK/United Kingdom
- FC001002/ARC_/Arthritis Research UK/United Kingdom
- 20466/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- C7893/A26233/CRUK_/Cancer Research UK/United Kingdom
- FC001130/ARC_/Arthritis Research UK/United Kingdom
- 17786/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

