Morphological and Taxonomic Properties of the Newly Isolated Cotonvirus japonicus, a New Lineage of the Subfamily Megavirinae
- PMID: 34191583
- PMCID: PMC8387033
- DOI: 10.1128/JVI.00919-21
Morphological and Taxonomic Properties of the Newly Isolated Cotonvirus japonicus, a New Lineage of the Subfamily Megavirinae
Abstract
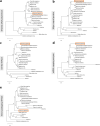
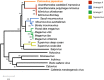
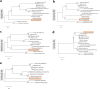
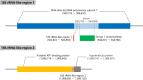
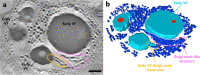
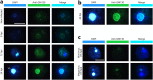
Since 2003, various viruses from the subfamily Megavirinae in the family Mimiviridae have been isolated worldwide, including icosahedral mimiviruses and tailed tupanviruses. To date, the evolutionary relationship between tailed and nontailed mimiviruses has not been elucidated. Here, we present the genomic and morphological features of a newly isolated giant virus, Cotonvirus japonicus (cotonvirus), belonging to the family Mimiviridae. It contains a linear double-stranded DNA molecule of 1.47 Mb, the largest among the reported viruses in the subfamily Megavirinae, excluding tupanviruses. Among its 1,306 predicted open reading frames, 1,149 (88.0%) were homologous to those of the family Mimiviridae. Several nucleocytoplasmic large DNA virus (NCLDV) core genes, aminoacyl-tRNA synthetase genes, and the host specificity of cotonvirus were highly similar to those of Mimiviridae lineages A, B, and C; however, lineage A was slightly closer to cotonvirus than the others were. Moreover, based on its genome size, the presence of two copies of 18S rRNA-like sequences, and the period of its infection cycle, cotonvirus is the most similar to the tupanviruses among the icosahedral mimiviruses. Interestingly, the cotonvirus utilizes Golgi apparatus-like vesicles for virion factory (VF) formation. Overall, we showed that cotonvirus is a novel lineage of the subfamily Megavirinae. Our findings support the diversity of icosahedral mimiviruses and provide mechanistic insights into the replication, VF formation, and evolution of the subfamily Megavirinae. IMPORTANCE We have isolated a new virus of an independent lineage belonging to the family Mimiviridae, subfamily Megavirinae, from the fresh water of a canal in Japan, named Cotonvirus. In a proteomic tree, this new nucleocytoplasmic large DNA virus (NCLDV) is phylogenetically placed at the root of three lineages of the subfamily Megavirinae-lineages A (mimivirus), B (moumouvirus), and C (megavirus). Multiple genomic and phenotypic features of cotonvirus are more similar to those of tupanviruses than to those of the A, B, or C lineages, and other genomic features, while the host specificity of cotonvirus is more similar to those of the latter than of the former. These results suggest that cotonvirus is a unique virus that has chimeric features of existing viruses of Megavirinae and uses Golgi apparatus-like vesicles of the host cells for virion factory (VF) formation. Thus, cotonvirus can provide novel insights into the evolution of mimiviruses and the underlying mechanisms of VF formation.
Keywords: Cotonvirus japonicus; family Mimiviridae; giant virus; isolation.
Figures



Similar articles
-
Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage.Genome Biol Evol. 2012;4(12):1324-30. doi: 10.1093/gbe/evs109. Genome Biol Evol. 2012. PMID: 23221609 Free PMC article.
-
Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae.Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17486-91. doi: 10.1073/pnas.1110889108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987820 Free PMC article.
-
Isolation and genomic characterization of a new mimivirus of lineage B from a Brazilian river.Arch Virol. 2020 Apr;165(4):853-863. doi: 10.1007/s00705-020-04542-5. Epub 2020 Feb 12. Arch Virol. 2020. PMID: 32052196
-
Mimivirus and its virophage.Annu Rev Genet. 2009;43:49-66. doi: 10.1146/annurev-genet-102108-134255. Annu Rev Genet. 2009. PMID: 19653859 Review.
-
Mimivirus: leading the way in the discovery of giant viruses of amoebae.Nat Rev Microbiol. 2017 Apr;15(4):243-254. doi: 10.1038/nrmicro.2016.197. Epub 2017 Feb 27. Nat Rev Microbiol. 2017. PMID: 28239153 Free PMC article. Review.
Cited by
-
Widespread Distribution and Evolution of Poxviral Entry-Fusion Complex Proteins in Giant Viruses.Microbiol Spectr. 2023 Mar 13;11(2):e0494422. doi: 10.1128/spectrum.04944-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36912656 Free PMC article.
-
Giant viruses of the Megavirinae subfamily possess biosynthetic pathways to produce rare bacterial-like sugars in a clade-specific manner.Microlife. 2022 Apr 6;3:uqac002. doi: 10.1093/femsml/uqac002. eCollection 2022. Microlife. 2022. PMID: 37223350 Free PMC article.
-
Complete genome sequence of Tornadovirus japonicus, a relative of Pacmanvirus, isolated from the Tamagawa River in Japan.Microbiol Resour Announc. 2024 Jul 18;13(7):e0026524. doi: 10.1128/mra.00265-24. Epub 2024 Jun 11. Microbiol Resour Announc. 2024. PMID: 38860801 Free PMC article.
-
Kinetic Analysis of Acanthamoeba castellanii Infected with Giant Viruses Quantitatively Revealed Process of Morphological and Behavioral Changes in Host Cells.Microbiol Spectr. 2021 Sep 3;9(1):e0036821. doi: 10.1128/Spectrum.00368-21. Epub 2021 Aug 25. Microbiol Spectr. 2021. PMID: 34431709 Free PMC article.
-
Establishment of a novel human T-cell leukemia virus type 1 infection model using cell-free virus.J Virol. 2024 Feb 20;98(2):e0186223. doi: 10.1128/jvi.01862-23. Epub 2024 Jan 31. J Virol. 2024. PMID: 38294250 Free PMC article.
References
-
- Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the Megavirus lineage. Genome Biol Evol 4:1324–1330. 10.1093/gbe/evs109. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

