Genome-wide expression and network analyses of mutants in key brassinosteroid signaling genes
- PMID: 34157989
- PMCID: PMC8220701
- DOI: 10.1186/s12864-021-07778-w
Genome-wide expression and network analyses of mutants in key brassinosteroid signaling genes
Abstract
Background: Brassinosteroid (BR) signaling regulates plant growth and development in concert with other signaling pathways. Although many genes have been identified that play a role in BR signaling, the biological and functional consequences of disrupting those key BR genes still require detailed investigation.
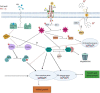
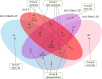
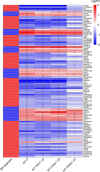
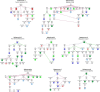
Results: Here we performed phenotypic and transcriptomic comparisons of A. thaliana lines carrying a loss-of-function mutation in BRI1 gene, bri1-5, that exhibits a dwarf phenotype and its three activation-tag suppressor lines that were able to partially revert the bri1-5 mutant phenotype to a WS2 phenotype, namely bri1-5/bri1-1D, bri1-5/brs1-1D, and bri1-5/bak1-1D. From the three investigated bri1-5 suppressors, bri1-5/bak1-1D was the most effective suppressor at the transcriptional level. All three bri1-5 suppressors showed altered expression of the genes in the abscisic acid (ABA signaling) pathway, indicating that ABA likely contributes to the partial recovery of the wild-type phenotype in these bri1-5 suppressors. Network analysis revealed crosstalk between BR and other phytohormone signaling pathways, suggesting that interference with one hormone signaling pathway affects other hormone signaling pathways. In addition, differential expression analysis suggested the existence of a strong negative feedback from BR signaling on BR biosynthesis and also predicted that BRS1, rather than being directly involved in signaling, might be responsible for providing an optimal environment for the interaction between BRI1 and its ligand.
Conclusions: Our study provides insights into the molecular mechanisms and functions of key brassinosteroid (BR) signaling genes, especially BRS1.
Keywords: Arabidopsis; Brassinosteroid signaling; Expression analysis; Network analysis; Systems biology.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling.PLoS Genet. 2012 Jan;8(1):e1002452. doi: 10.1371/journal.pgen.1002452. Epub 2012 Jan 12. PLoS Genet. 2012. PMID: 22253607 Free PMC article.
-
BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2001 May 8;98(10):5916-21. doi: 10.1073/pnas.091065998. Epub 2001 Apr 24. Proc Natl Acad Sci U S A. 2001. PMID: 11320207 Free PMC article.
-
14-3-3 proteins contribute to leaf and root development via brassinosteroid insensitive 1 in Arabidopsis thaliana.Genes Genomics. 2020 Mar;42(3):347-354. doi: 10.1007/s13258-019-00909-4. Epub 2020 Jan 4. Genes Genomics. 2020. PMID: 31902106
-
Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1.J Integr Plant Biol. 2013 Dec;55(12):1198-211. doi: 10.1111/jipb.12081. Epub 2013 Sep 9. J Integr Plant Biol. 2013. PMID: 23718739 Review.
-
BAK1 directly regulates brassinosteroid perception and BRI1 activation.J Integr Plant Biol. 2013 Dec;55(12):1264-70. doi: 10.1111/jipb.12122. J Integr Plant Biol. 2013. PMID: 24308570 Review.
Cited by
-
The cap-binding complex modulates ABA-responsive transcript splicing during germination in barley (Hordeum vulgare).Sci Rep. 2024 Aug 7;14(1):18278. doi: 10.1038/s41598-024-69373-9. Sci Rep. 2024. PMID: 39107424 Free PMC article.
-
Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root.Science. 2023 Mar 31;379(6639):eadf4721. doi: 10.1126/science.adf4721. Epub 2023 Mar 31. Science. 2023. PMID: 36996230 Free PMC article.
-
Excavation of Genes Responsive to Brassinosteroids by Transcriptome Sequencing in Adiantum flabellulatum Gametophytes.Genes (Basel). 2022 Jun 14;13(6):1061. doi: 10.3390/genes13061061. Genes (Basel). 2022. PMID: 35741824 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

