Endogenous modulators of neurotrophin signaling: Landscape of the transient ATP-NGF interactions
- PMID: 34136093
- PMCID: PMC8164016
- DOI: 10.1016/j.csbj.2021.05.009
Endogenous modulators of neurotrophin signaling: Landscape of the transient ATP-NGF interactions
Abstract
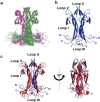
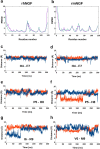
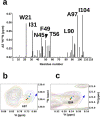
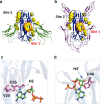
The Nerve Growth Factor (NGF) neurotrophin acts in the maintenance and growth of neuronal populations. Despite the detailed knowledge of NGF's role in neuron physiology, the structural and mechanistic determinants of NGF bioactivity modulated by essential endogenous ligands are still lacking. We present the results of an integrated structural and advanced computational approach to characterize the extracellular ATP-NGF interaction. We mapped by NMR the interacting surface and ATP orientation on NGF and revealed the functional role of this interaction in the binding to TrkA and p75NTR receptors by SPR. The role of divalent ions was explored in conjunction with ATP. Our results pinpoint ATP as a likely transient molecular modulator of NGF signaling, in health and disease states.
Keywords: ARIA, Ambiguous Restraints for Iterative Assignment; ATP modulation; BDNF, Brain Derived Neurotrophic Factor; CARA, Computer Aided Resonance Assignment; CS-E, Chrondroitin Sulfate E; CSP, Chemical Shift Perturbation; DSF, Differential Scanning Fluorimetry; EI-MS, Electron Ionization Mass Spectrometry; Endogenous ligands; FGF2, Fibroblast Growth Factor 2; FT-IR, Fourier Transform Infrared Spectroscopy; HBD, Heparin Binding Domain; HSQC, Heteronuclear Single Quantum Coherence; ITC, Isothermal Titration Calorimetry; MALDI-TOF MS, Matrix Assisted Laser Desorption Ionization-Time Of Flight Mass Spectrometry; MD, Molecular Dynamics; MS, Mass Spectrometry; NGF interactions; NGF, Nerve Growth Factor; NMR, Nuclear Magnetic Resonance; NOE, Nuclear Overhouser Effect; NOESY, Nuclear Overhauser Effect Spectroscopy; NT, NeuroTrophin; Neurotrophins; P20, Polysorbate 20; PME, Particle Mesh Ewald; RMSD, Root Mean Square Deviation; SAR, Structure-Activity Relationship; SPR, Surface Plasmon Resonance; STD, Saturation-Transfer Difference; TrkA, Tyrosine Kinase Receptor A; TrkA, p75NTR receptors; p75NTR, p75 NeuroTrophin Receptor; proNGF, proNGF – NGF precursor; rh-proNGF, recombinant human proNGF – NGF precursor; rhNGF, recombinant human NGF; rmNGF, recombinant mouse NGF.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Small Endogenous Ligands Modulation of Nerve Growth Factor Bioactivity: A Structural Biology Overview.Cells. 2021 Dec 8;10(12):3462. doi: 10.3390/cells10123462. Cells. 2021. PMID: 34943971 Free PMC article. Review.
-
Distinct conformational changes occur within the intrinsically unstructured pro-domain of pro-Nerve Growth Factor in the presence of ATP and Mg2.Protein Sci. 2023 Feb;32(2):e4563. doi: 10.1002/pro.4563. Protein Sci. 2023. PMID: 36605018 Free PMC article.
-
Using surface plasmon resonance spectroscopy to characterize the inhibition of NGF-p75(NTR) and proNGF-p75(NTR) interactions by small molecule inhibitors.Pharmacol Res. 2016 Jan;103:292-9. doi: 10.1016/j.phrs.2015.12.005. Epub 2015 Dec 7. Pharmacol Res. 2016. PMID: 26675716
-
Type I interferons up-regulate the expression and signalling of p75 NTR/TrkA receptor complex in differentiated human SH-SY5Y neuroblastoma cells.Neuropharmacology. 2014 Apr;79:321-34. doi: 10.1016/j.neuropharm.2013.12.002. Epub 2013 Dec 11. Neuropharmacology. 2014. PMID: 24333329
-
ProNGF and Neurodegeneration in Alzheimer's Disease.Front Neurosci. 2019 Feb 22;13:129. doi: 10.3389/fnins.2019.00129. eCollection 2019. Front Neurosci. 2019. PMID: 30853882 Free PMC article. Review.
Cited by
-
Small Endogenous Ligands Modulation of Nerve Growth Factor Bioactivity: A Structural Biology Overview.Cells. 2021 Dec 8;10(12):3462. doi: 10.3390/cells10123462. Cells. 2021. PMID: 34943971 Free PMC article. Review.
-
Distinct conformational changes occur within the intrinsically unstructured pro-domain of pro-Nerve Growth Factor in the presence of ATP and Mg2.Protein Sci. 2023 Feb;32(2):e4563. doi: 10.1002/pro.4563. Protein Sci. 2023. PMID: 36605018 Free PMC article.
References
-
- Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237(4819):1154–1162. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
