A bioinformatics approach for identifying potential molecular mechanisms and key genes involved in COVID-19 associated cardiac remodeling
- PMID: 34131597
- PMCID: PMC8192842
- DOI: 10.1016/j.genrep.2021.101246
A bioinformatics approach for identifying potential molecular mechanisms and key genes involved in COVID-19 associated cardiac remodeling
Abstract
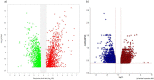
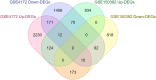
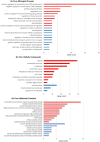
In 2019 coronavirus disease (COVID-19), whose main complication is respiratory involvement, different organs may also be affected in severe cases. However, COVID-19 associated cardiovascular manifestations are limited at present. The main purpose of this study was to identify potential candidate genes involved in COVID-19-associated heart damage by bioinformatics analysis. Differently expressed genes (DEGs) were identified using transcriptome profiles (GSE150392 and GSE4172) downloaded from the GEO database. After gene and pathway enrichment analyses, PPI network visualization, module analyses, and hub gene extraction were performed using Cytoscape software. A total of 228 (136 up and 92 downregulated) overlapping DEGs were identified at these two microarray datasets. Finally, the top hub genes (FGF2, JUN, TLR4, and VEGFA) were screened out as the critical genes among the DEGs from the PPI network. Identification of critical genes and mechanisms in any disease can lead us to better diagnosis and targeted therapy. Our findings identified core genes shared by inflammatory cardiomyopathy and SARS-CoV-2. The findings of the current study support the idea that these key genes can be used in understanding and managing the long-term cardiovascular effects of COVID-19.
Keywords: ACE2, angiotensin-converting enzyme 2; Bioinformatics analysis; COVID-19; COVID-19, the coronavirus disease 2019; Cardiac remodeling; DEGs, differentially expressed genes; Differential expression; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genome; PPI, the protein-protein interaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; STRING, the search tool for the retrieval of interacting genes.
© 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
The author declares that there is no conflict of interest with any financial organization or corporation or individual that can inappropriately influence this work.
Figures
Similar articles
-
Identification of the Hub Genes and the Signaling Pathways in Human iPSC-Cardiomyocytes Infected by SARS-CoV-2.Biochem Genet. 2022 Dec;60(6):2052-2068. doi: 10.1007/s10528-022-10206-7. Epub 2022 Mar 2. Biochem Genet. 2022. PMID: 35235083 Free PMC article.
-
Identifying MMP14 and COL12A1 as a potential combination of prognostic biomarkers in pancreatic ductal adenocarcinoma using integrated bioinformatics analysis.PeerJ. 2020 Nov 23;8:e10419. doi: 10.7717/peerj.10419. eCollection 2020. PeerJ. 2020. PMID: 33282565 Free PMC article.
-
Blood transcriptome analysis revealed the crosstalk between COVID-19 and HIV.Front Immunol. 2022 Oct 28;13:1008653. doi: 10.3389/fimmu.2022.1008653. eCollection 2022. Front Immunol. 2022. PMID: 36389792 Free PMC article.
-
Identification of Crucial Genes and Key Functions in Type 2 Diabetic Hearts by Bioinformatic Analysis.Front Endocrinol (Lausanne). 2022 Feb 15;13:801260. doi: 10.3389/fendo.2022.801260. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35242109 Free PMC article.
-
Construction of miRNA-mRNA network for the identification of key biological markers and their associated pathways in IgA nephropathy by employing the integrated bioinformatics analysis.Saudi J Biol Sci. 2021 Sep;28(9):4938-4945. doi: 10.1016/j.sjbs.2021.06.079. Epub 2021 Jul 1. Saudi J Biol Sci. 2021. PMID: 34466069 Free PMC article.
Cited by
-
Identification of critical genes and molecular pathways in COVID-19 myocarditis and constructing gene regulatory networks by bioinformatic analysis.PLoS One. 2022 Jun 24;17(6):e0269386. doi: 10.1371/journal.pone.0269386. eCollection 2022. PLoS One. 2022. PMID: 35749386 Free PMC article.
-
Integrated Bioinformatics Analysis to Identify Alternative Therapeutic Targets for Alzheimer's Disease: Insights from a Synaptic Machinery Perspective.J Mol Neurosci. 2022 Feb;72(2):273-286. doi: 10.1007/s12031-021-01893-9. Epub 2021 Aug 19. J Mol Neurosci. 2022. PMID: 34414562
-
A nine-hub-gene signature of metabolic syndrome identified using machine learning algorithms and integrated bioinformatics.Bioengineered. 2021 Dec;12(1):5727-5738. doi: 10.1080/21655979.2021.1968249. Bioengineered. 2021. PMID: 34516309 Free PMC article.
-
A systems biology approach for investigating significantly expressed genes among COVID-19, hepatocellular carcinoma, and chronic hepatitis B.Egypt J Med Hum Genet. 2022;23(1):146. doi: 10.1186/s43042-022-00360-3. Epub 2022 Oct 20. Egypt J Med Hum Genet. 2022. PMID: 37521843 Free PMC article.
-
In-silico discovery of common molecular signatures for which SARS-CoV-2 infections and lung diseases stimulate each other, and drug repurposing.PLoS One. 2024 Jul 18;19(7):e0304425. doi: 10.1371/journal.pone.0304425. eCollection 2024. PLoS One. 2024. PMID: 39024368 Free PMC article.
References
-
- Bahrami S., Drabløs F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016;62:37–49. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
