Structure-mechanics statistical learning unravels the linkage between local rigidity and global flexibility in nucleic acids
- PMID: 34122953
- PMCID: PMC8159235
- DOI: 10.1039/d0sc00480d
Structure-mechanics statistical learning unravels the linkage between local rigidity and global flexibility in nucleic acids
Abstract
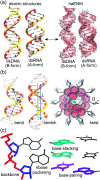
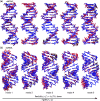
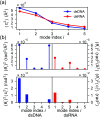
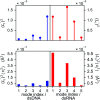
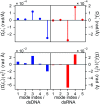
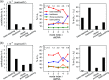
The mechanical properties of nucleic acids underlie biological processes ranging from genome packaging to gene expression, but tracing their molecular origin has been difficult due to the structural and chemical complexity. We posit that concepts from machine learning can help to tackle this long-standing challenge. Here, we demonstrate the feasibility and advantage of this strategy through developing a structure-mechanics statistical learning scheme to elucidate how local rigidity in double-stranded (ds)DNA and dsRNA may lead to their global flexibility in bend, stretch, and twist. Specifically, the mechanical parameters in a heavy-atom elastic network model are computed from the trajectory data of all-atom molecular dynamics simulation. The results show that the inter-atomic springs for backbone and ribose puckering in dsRNA are stronger than those in dsDNA, but are similar in strengths for base-stacking and base-pairing. Our analysis shows that the experimental observation of dsDNA being easier to bend but harder to stretch than dsRNA comes mostly from the respective B- and A-form topologies. The computationally resolved composition of local rigidity indicates that the flexibility of both nucleic acids is mostly due to base-stacking. But for properties like twist-stretch coupling, backbone springs are shown to play a major role instead. The quantitative connection between local rigidity and global flexibility sets foundation for understanding how local binding and chemical modification of genetic materials effectuate longer-ranged regulatory signals.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare statement.
Figures

Similar articles
-
Understanding the Relative Flexibility of RNA and DNA Duplexes: Stretching and Twist-Stretch Coupling.Biophys J. 2017 Mar 28;112(6):1094-1104. doi: 10.1016/j.bpj.2017.02.022. Biophys J. 2017. PMID: 28355538 Free PMC article.
-
Understanding the mechanical response of double-stranded DNA and RNA under constant stretching forces using all-atom molecular dynamics.Proc Natl Acad Sci U S A. 2017 Jul 3;114(27):7049-7054. doi: 10.1073/pnas.1705642114. Epub 2017 Jun 20. Proc Natl Acad Sci U S A. 2017. PMID: 28634300 Free PMC article.
-
Structural Flexibility of DNA-RNA Hybrid Duplex: Stretching and Twist-Stretch Coupling.Biophys J. 2019 Jul 9;117(1):74-86. doi: 10.1016/j.bpj.2019.05.018. Epub 2019 May 23. Biophys J. 2019. PMID: 31164196 Free PMC article.
-
A Tour de Force on the Double Helix: Exploiting DNA Mechanics To Study DNA-Based Molecular Machines.Biochemistry. 2019 Nov 26;58(47):4667-4676. doi: 10.1021/acs.biochem.9b00346. Epub 2019 Jun 28. Biochemistry. 2019. PMID: 31251042 Free PMC article. Review.
-
Aromatic Base Stacking in DNA: From ab initio Calculations to Molecular Dynamics Simulations.J Biomol Struct Dyn. 2000;17 Suppl 1:1-24. doi: 10.1080/07391102.2000.10506597. J Biomol Struct Dyn. 2000. PMID: 22607400 Review.
Cited by
-
Mechanical Flexibility of DNA: A Quintessential Tool for DNA Nanotechnology.Sensors (Basel). 2020 Dec 8;20(24):7019. doi: 10.3390/s20247019. Sensors (Basel). 2020. PMID: 33302459 Free PMC article. Review.
-
Structure-mechanics statistical learning uncovers mechanical relay in proteins.Chem Sci. 2022 Jan 19;13(13):3688-3696. doi: 10.1039/d1sc06184d. eCollection 2022 Mar 30. Chem Sci. 2022. PMID: 35432911 Free PMC article.
-
Mechanical couplings of protein backbone and side chains exhibit scale-free network properties and specific hotspots for function.Comput Struct Biotechnol J. 2021 Sep 8;19:5309-5320. doi: 10.1016/j.csbj.2021.09.004. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34765086 Free PMC article.
-
Mechanical codes of chemical-scale specificity in DNA motifs.Chem Sci. 2023 Aug 29;14(37):10155-10166. doi: 10.1039/d3sc01671d. eCollection 2023 Sep 27. Chem Sci. 2023. PMID: 37772098 Free PMC article.
-
Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.Nucleic Acids Res. 2023 Nov 27;51(21):11927-11940. doi: 10.1093/nar/gkad910. Nucleic Acids Res. 2023. PMID: 37870446 Free PMC article.
References
LinkOut - more resources
Full Text Sources

