Insight Into Spinocerebellar Ataxia Type 31 (SCA31) From Drosophila Model
- PMID: 34113230
- PMCID: PMC8185138
- DOI: 10.3389/fnins.2021.648133
Insight Into Spinocerebellar Ataxia Type 31 (SCA31) From Drosophila Model
Abstract
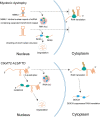
Spinocerebellar ataxia type 31 (SCA31) is a progressive neurodegenerative disease characterized by degeneration of Purkinje cells in the cerebellum. Its genetic cause is a 2.5- to 3.8-kb-long complex pentanucleotide repeat insertion containing (TGGAA)n, (TAGAA)n, (TAAAA)n, and (TAAAATAGAA)n located in an intron shared by two different genes: brain expressed associated with NEDD4-1 (BEAN1) and thymidine kinase 2 (TK2). Among these repeat sequences, (TGGAA)n repeat was the only sequence segregating with SCA31, which strongly suggests its pathogenicity. In SCA31 patient brains, the mutant BEAN1 transcript containing expanded UGGAA repeats (UGGAAexp) was found to form abnormal RNA structures called RNA foci in cerebellar Purkinje cell nuclei. In addition, the deposition of pentapeptide repeat (PPR) proteins, poly(Trp-Asn-Gly-Met-Glu), translated from UGGAAexp RNA, was detected in the cytoplasm of Purkinje cells. To uncover the pathogenesis of UGGAAexp in SCA31, we generated Drosophila models of SCA31 expressing UGGAAexp RNA. The toxicity of UGGAAexp depended on its length and expression level, which was accompanied by the accumulation of RNA foci and translation of repeat-associated PPR proteins in Drosophila, consistent with the observation in SCA31 patient brains. We also revealed that TDP-43, FUS, and hnRNPA2B1, motor neuron disease-linked RNA-binding proteins bound to UGGAAexp RNA, act as RNA chaperones to regulate the formation of RNA foci and repeat-associated translation. Further research on the role of RNA-binding proteins as RNA chaperones may also provide a novel therapeutic strategy for other microsatellite repeat expansion diseases besides SCA31.
Keywords: RAN translation; RBP; RNA chaperone; RNA foci; SCA31; TDP-43; microsatellite repeat.
Copyright © 2021 Ishiguro, Nagai and Ishikawa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Molecular Mechanisms and Future Therapeutics for Spinocerebellar Ataxia Type 31 (SCA31).Neurotherapeutics. 2019 Oct;16(4):1106-1114. doi: 10.1007/s13311-019-00804-6. Neurotherapeutics. 2019. PMID: 31755042 Free PMC article. Review.
-
Abnormal RNA structures (RNA foci) containing a penta-nucleotide repeat (UGGAA)n in the Purkinje cell nucleus is associated with spinocerebellar ataxia type 31 pathogenesis.Neuropathology. 2013 Dec;33(6):600-11. doi: 10.1111/neup.12032. Epub 2013 Apr 22. Neuropathology. 2013. PMID: 23607545
-
Regulatory Role of RNA Chaperone TDP-43 for RNA Misfolding and Repeat-Associated Translation in SCA31.Neuron. 2017 Apr 5;94(1):108-124.e7. doi: 10.1016/j.neuron.2017.02.046. Epub 2017 Mar 23. Neuron. 2017. PMID: 28343865 Free PMC article.
-
Spinocerebellar ataxia type 31 (SCA31).J Hum Genet. 2023 Mar;68(3):153-156. doi: 10.1038/s10038-022-01091-4. Epub 2022 Nov 1. J Hum Genet. 2023. PMID: 36319738 Free PMC article. Review.
-
[Spinocerebellar ataxia type 31].Rinsho Shinkeigaku. 2010 Nov;50(11):985-7. doi: 10.5692/clinicalneurol.50.985. Rinsho Shinkeigaku. 2010. PMID: 21921537 Review. Japanese.
Cited by
-
Rapid and comprehensive diagnostic method for repeat expansion diseases using nanopore sequencing.NPJ Genom Med. 2022 Oct 26;7(1):62. doi: 10.1038/s41525-022-00331-y. NPJ Genom Med. 2022. PMID: 36289212 Free PMC article.
-
RNA Modifications and RNA Metabolism in Neurological Disease Pathogenesis.Int J Mol Sci. 2021 Nov 1;22(21):11870. doi: 10.3390/ijms222111870. Int J Mol Sci. 2021. PMID: 34769301 Free PMC article. Review.
-
Elevated BEAN1 expression correlates with poor prognosis, immune evasion, and chemotherapy resistance in rectal adenocarcinoma.Discov Oncol. 2024 Sep 14;15(1):446. doi: 10.1007/s12672-024-01321-5. Discov Oncol. 2024. PMID: 39276259 Free PMC article.
-
Genetic modifiers of repeat expansion disorders.Emerg Top Life Sci. 2023 Dec 14;7(3):325-337. doi: 10.1042/ETLS20230015. Emerg Top Life Sci. 2023. PMID: 37861103 Free PMC article.
-
Mechanistic and Therapeutic Insights into Ataxic Disorders with Pentanucleotide Expansions.Cells. 2022 May 6;11(9):1567. doi: 10.3390/cells11091567. Cells. 2022. PMID: 35563872 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

