A new molecular classification to drive precision treatment strategies in primary Sjögren's syndrome
- PMID: 34112769
- PMCID: PMC8192578
- DOI: 10.1038/s41467-021-23472-7
A new molecular classification to drive precision treatment strategies in primary Sjögren's syndrome
Abstract
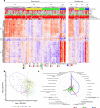
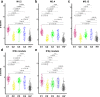
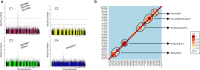
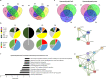
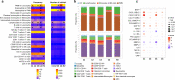
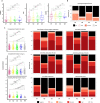
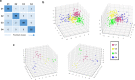
There is currently no approved treatment for primary Sjögren's syndrome, a disease that primarily affects adult women. The difficulty in developing effective therapies is -in part- because of the heterogeneity in the clinical manifestation and pathophysiology of the disease. Finding common molecular signatures among patient subgroups could improve our understanding of disease etiology, and facilitate the development of targeted therapeutics. Here, we report, in a cross-sectional cohort, a molecular classification scheme for Sjögren's syndrome patients based on the multi-omic profiling of whole blood samples from a European cohort of over 300 patients, and a similar number of age and gender-matched healthy volunteers. Using transcriptomic, genomic, epigenetic, cytokine expression and flow cytometry data, combined with clinical parameters, we identify four groups of patients with distinct patterns of immune dysregulation. The biomarkers we identify can be used by machine learning classifiers to sort future patients into subgroups, allowing the re-evaluation of response to treatments in clinical trials.
Conflict of interest statement
While engaged in the research project, R.L., F.M., and Z.M. were regular employees of Bayer A.G. At present, R.L. and Z.M. are regular employees of Nuvisan ICB GmbH, a company providing contract research services. P.S., S.H., S.C.G., L.X., M.G., P.M., and L.L. were regular employees of Institut de Recherches Internationales Servier at the time of the research project. B.C., C.B., and E.D. were phD students financed by Institut de Recherches Internationales Servier when they contributed to the research project. All other authors confirmed signing the ICMJE form for Disclosure of Potential Conflicts of Interest and none of them have any conflict of interest related to this work.
Figures

Similar articles
-
The Transcriptomic Profile of Monocytes from Patients With Sjögren's Syndrome Is Associated With Inflammatory Parameters and Is Mimicked by Circulating Mediators.Front Immunol. 2021 Aug 3;12:701656. doi: 10.3389/fimmu.2021.701656. eCollection 2021. Front Immunol. 2021. PMID: 34413853 Free PMC article.
-
Cytokine concentrations in stimulated whole saliva among patients with primary Sjögren's syndrome, secondary Sjögren's syndrome, and patients with primary Sjögren's syndrome receiving varying doses of interferon for symptomatic treatment of the condition: a preliminary study.Clin Oral Investig. 2001 Jun;5(2):133-5. doi: 10.1007/s007840100104. Clin Oral Investig. 2001. PMID: 11480812 Clinical Trial.
-
Integrated Bioinformatics and Validation Reveal Potential Biomarkers Associated With Progression of Primary Sjögren's Syndrome.Front Immunol. 2021 Jul 23;12:697157. doi: 10.3389/fimmu.2021.697157. eCollection 2021. Front Immunol. 2021. PMID: 34367157 Free PMC article.
-
Sjögren's syndrome--a plethora of clinical and immunological phenotypes with a complex genetic background.Ann N Y Acad Sci. 2007 Jun;1108:433-47. doi: 10.1196/annals.1422.046. Ann N Y Acad Sci. 2007. PMID: 17894008 Review.
-
[Autoantibodies in patients with Sjögren's syndrome and their clinical significance].Nihon Rinsho. 1995 Oct;53(10):2422-8. Nihon Rinsho. 1995. PMID: 8531349 Review. Japanese.
Cited by
-
Dysregulated long non-coding RNA in Sjögren's disease impacts both interferon and adaptive immune responses.RMD Open. 2022 Nov;8(2):e002672. doi: 10.1136/rmdopen-2022-002672. RMD Open. 2022. PMID: 36456101 Free PMC article.
-
Feature-specific quantile normalization and feature-specific mean-variance normalization deliver robust bi-directional classification and feature selection performance between microarray and RNAseq data.BMC Bioinformatics. 2024 Mar 29;25(1):136. doi: 10.1186/s12859-024-05759-w. BMC Bioinformatics. 2024. PMID: 38549046 Free PMC article.
-
Expression and regulatory characteristics of peripheral blood immune cells in primary Sjögren's syndrome patients using single-cell transcriptomic.iScience. 2022 Nov 4;25(12):105509. doi: 10.1016/j.isci.2022.105509. eCollection 2022 Dec 22. iScience. 2022. PMID: 36425764 Free PMC article.
-
Novel multiclass classification machine learning approach for the early-stage classification of systemic autoimmune rheumatic diseases.Lupus Sci Med. 2024 Jan 31;11(1):e001125. doi: 10.1136/lupus-2023-001125. Lupus Sci Med. 2024. PMID: 38302133 Free PMC article.
-
CCL5 Release by CCR9+ CD8 T Cells: A Potential Contributor to Immunopathology of Primary Sjögren's Syndrome.Front Immunol. 2022 Jun 1;13:887972. doi: 10.3389/fimmu.2022.887972. eCollection 2022. Front Immunol. 2022. PMID: 35720379 Free PMC article.
References
-
- Barturen, G. et al. Integrative analysis reveals a molecular stratification of systemic autoimmune diseases. Arthritis Rheumatol. (2020) 10.1002/art.41610 https://doi:10.17881/th9v-xt85. - PubMed

