Depalmitoylation rewires FLT3-ITD signaling and exacerbates leukemia progression
- PMID: 34111291
- PMCID: PMC8832469
- DOI: 10.1182/blood.2021011582
Depalmitoylation rewires FLT3-ITD signaling and exacerbates leukemia progression
Abstract
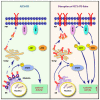
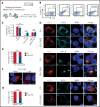
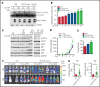
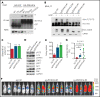
Internal tandem duplication within FLT3 (FLT3-ITD) is one of the most frequent mutations in acute myeloid leukemia (AML) and correlates with a poor prognosis. Whereas the FLT3 receptor tyrosine kinase is activated at the plasma membrane to transduce PI3K/AKT and RAS/MAPK signaling, FLT3-ITD resides in the endoplasmic reticulum and triggers constitutive STAT5 phosphorylation. Mechanisms underlying this aberrant FLT3-ITD subcellular localization or its impact on leukemogenesis remain poorly established. In this study, we discovered that FLT3-ITD is S-palmitoylated by the palmitoyl acyltransferase ZDHHC6. Disruption of palmitoylation redirected FLT3-ITD to the plasma membrane and rewired its downstream signaling by activating AKT and extracellular signal-regulated kinase pathways in addition to STAT5. Consequently, abrogation of palmitoylation increased FLT3-ITD-mediated progression of leukemia in xenotransplant-recipient mouse models. We further demonstrate that FLT3 proteins were palmitoylated in primary human AML cells. ZDHHC6-mediated palmitoylation restrained FLT3-ITD surface expression, signaling, and colonogenic growth of primary FLT3-ITD+ AML. More important, pharmacological inhibition of FLT3-ITD depalmitoylation synergized with the US Food and Drug Administration-approved FLT3 kinase inhibitor gilteritinib in abrogating the growth of primary FLT3-ITD+ AML cells. These findings provide novel insights into lipid-dependent compartmentalization of FLT3-ITD signaling in AML and suggest targeting depalmitoylation as a new therapeutic strategy to treat FLT3-ITD+ leukemias.
© 2021 by The American Society of Hematology.
Figures



Comment in
-
Fattening up FLT3-ITD for the kill.Blood. 2021 Dec 2;138(22):2158-2159. doi: 10.1182/blood.2021013182. Blood. 2021. PMID: 34854880 No abstract available.
Similar articles
-
Modulation of FLT3-ITD Localization and Targeting of Distinct Downstream Signaling Pathways as Potential Strategies to Overcome FLT3-Inhibitor Resistance.Cells. 2021 Nov 3;10(11):2992. doi: 10.3390/cells10112992. Cells. 2021. PMID: 34831215 Free PMC article.
-
Pim-1 kinase phosphorylates and stabilizes 130 kDa FLT3 and promotes aberrant STAT5 signaling in acute myeloid leukemia with FLT3 internal tandem duplication.PLoS One. 2013 Sep 5;8(9):e74653. doi: 10.1371/journal.pone.0074653. eCollection 2013. PLoS One. 2013. PMID: 24040307 Free PMC article.
-
Survivin mediates aberrant hematopoietic progenitor cell proliferation and acute leukemia in mice induced by internal tandem duplication of Flt3.Blood. 2009 Jul 9;114(2):394-403. doi: 10.1182/blood-2008-11-188714. Epub 2009 May 1. Blood. 2009. PMID: 19411632 Free PMC article.
-
Targeting on glycosylation of mutant FLT3 in acute myeloid leukemia.Hematology. 2019 Dec;24(1):651-660. doi: 10.1080/16078454.2019.1666219. Hematology. 2019. PMID: 31533545 Review.
-
Quizartinib (AC220): a promising option for acute myeloid leukemia.Drug Des Devel Ther. 2019 Apr 8;13:1117-1125. doi: 10.2147/DDDT.S198950. eCollection 2019. Drug Des Devel Ther. 2019. PMID: 31114157 Free PMC article. Review.
Cited by
-
Novel Insights into the Role of Kras in Myeloid Differentiation: Engaging with Wnt/β-Catenin Signaling.Cells. 2023 Jan 14;12(2):322. doi: 10.3390/cells12020322. Cells. 2023. PMID: 36672256 Free PMC article. Review.
-
N6-methyladenosine RNA modifications: a potential therapeutic target for AML.Ann Hematol. 2024 Aug;103(8):2601-2612. doi: 10.1007/s00277-023-05302-6. Epub 2023 Aug 7. Ann Hematol. 2024. PMID: 37548690 Review.
-
Palmitoyltransferase ZDHHC3 is essential for the oncogenic activity of PML/RARα in acute promyelocytic leukemia.Acta Pharmacol Sin. 2025 Feb;46(2):462-473. doi: 10.1038/s41401-024-01371-z. Epub 2024 Sep 3. Acta Pharmacol Sin. 2025. PMID: 39227737
-
Atractylenolide-1 Targets FLT3 to Regulate PI3K/AKT/HIF1-α Pathway to Inhibit Osteogenic Differentiation of Human Valve Interstitial Cells.Front Pharmacol. 2022 Apr 25;13:899775. doi: 10.3389/fphar.2022.899775. eCollection 2022. Front Pharmacol. 2022. PMID: 35571096 Free PMC article.
-
Protein lipidation in health and disease: molecular basis, physiological function and pathological implication.Signal Transduct Target Ther. 2024 Mar 15;9(1):60. doi: 10.1038/s41392-024-01759-7. Signal Transduct Target Ther. 2024. PMID: 38485938 Free PMC article. Review.
References
-
- Port M, Böttcher M, Thol F, et al. . Prognostic significance of FLT3 internal tandem duplication, nucleophosmin 1, and CEBPA gene mutations for acute myeloid leukemia patients with normal karyotype and younger than 60 years - a systematic review and meta-analysis. Ann Hematol. 2014;93(8):1279-1286. - PubMed
-
- Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S.. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters--an analysis of 3082 patients. Blood. 2008;111(5):2527-2537. - PubMed
-
- Hayakawa F, Towatari M, Kiyoi H, et al. . Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19(5):624-631. - PubMed
-
- Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W.. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003; 9(6):2140-2150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

