De novo activated transcription of inserted foreign coding sequences is inheritable in the plant genome
- PMID: 34111139
- PMCID: PMC8191969
- DOI: 10.1371/journal.pone.0252674
De novo activated transcription of inserted foreign coding sequences is inheritable in the plant genome
Abstract
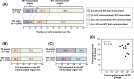
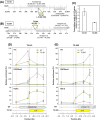
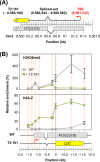
The manner in which inserted foreign coding sequences become transcriptionally activated and fixed in the plant genome is poorly understood. To examine such processes of gene evolution, we performed an artificial evolutionary experiment in Arabidopsis thaliana. As a model of gene-birth events, we introduced a promoterless coding sequence of the firefly luciferase (LUC) gene and established 386 T2-generation transgenic lines. Among them, we determined the individual LUC insertion loci in 76 lines and found that one-third of them were transcribed de novo even in the intergenic or inherently unexpressed regions. In the transcribed lines, transcription-related chromatin marks were detected across the newly activated transcribed regions. These results agreed with our previous findings in A. thaliana cultured cells under a similar experimental scheme. A comparison of the results of the T2-plant and cultured cell experiments revealed that the de novo-activated transcription concomitant with local chromatin remodelling was inheritable. During one-generation inheritance, it seems likely that the transcription activities of the LUC inserts trapped by the endogenous genes/transcripts became stronger, while those of de novo transcription in the intergenic/untranscribed regions became weaker. These findings may offer a clue for the elucidation of the mechanism by which inserted foreign coding sequences become transcriptionally activated and fixed in the plant genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Kozak Sequence Acts as a Negative Regulator for De Novo Transcription Initiation of Newborn Coding Sequences in the Plant Genome.Mol Biol Evol. 2021 Jun 25;38(7):2791-2803. doi: 10.1093/molbev/msab069. Mol Biol Evol. 2021. PMID: 33705557 Free PMC article.
-
Cryptic promoter activation occurs by at least two different mechanisms in the Arabidopsis genome.Plant J. 2021 Oct;108(1):29-39. doi: 10.1111/tpj.15420. Epub 2021 Aug 2. Plant J. 2021. PMID: 34252235
-
The coding sequence of firefly luciferase reporter gene affects specific hyperexpression in Arabidopsis thaliana cpl1 mutant.Plant Signal Behav. 2017 Aug 3;12(8):e1346767. doi: 10.1080/15592324.2017.1346767. Epub 2017 Jul 10. Plant Signal Behav. 2017. PMID: 28692335 Free PMC article.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
Cited by
-
Kozak Sequence Acts as a Negative Regulator for De Novo Transcription Initiation of Newborn Coding Sequences in the Plant Genome.Mol Biol Evol. 2021 Jun 25;38(7):2791-2803. doi: 10.1093/molbev/msab069. Mol Biol Evol. 2021. PMID: 33705557 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

