Synergistic roles for human U1 snRNA stem-loops in pre-mRNA splicing
- PMID: 34105434
- PMCID: PMC8632089
- DOI: 10.1080/15476286.2021.1932360
Synergistic roles for human U1 snRNA stem-loops in pre-mRNA splicing
Abstract
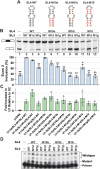
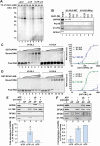
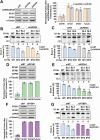
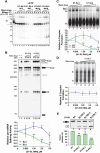
During spliceosome assembly, interactions that bring the 5' and 3' ends of an intron in proximity are critical for the production of mature mRNA. Here, we report synergistic roles for the stem-loops 3 (SL3) and 4 (SL4) of the human U1 small nuclear RNA (snRNA) in maintaining the optimal U1 snRNP function, and formation of cross-intron contact with the U2 snRNP. We find that SL3 and SL4 bind distinct spliceosomal proteins and combining a U1 snRNA activity assay with siRNA-mediated knockdown, we demonstrate that SL3 and SL4 act through the RNA helicase UAP56 and the U2 protein SF3A1, respectively. In vitro analysis using UV crosslinking and splicing assays indicated that SL3 likely promotes the SL4-SF3A1 interaction leading to enhancement of A complex formation and pre-mRNA splicing. Overall, these results highlight the vital role of the distinct contacts of SL3 and SL4 in bridging the pre-mRNA bound U1 and U2 snRNPs during the early steps of human spliceosome assembly.
Keywords: A complex; DDX39A; DDX39B; E complex; SF3A1; Spliceosome; Stem-loop; U1 snRNA; U1 snRNP; U2 snRNP; UAP56; URH49.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Principles and correction of 5'-splice site selection.RNA Biol. 2022 Jan;19(1):943-960. doi: 10.1080/15476286.2022.2100971. RNA Biol. 2022. PMID: 35866748 Free PMC article. Review.
-
Stem-loop 4 of U1 snRNA is essential for splicing and interacts with the U2 snRNP-specific SF3A1 protein during spliceosome assembly.Genes Dev. 2014 Nov 15;28(22):2518-31. doi: 10.1101/gad.248625.114. Genes Dev. 2014. PMID: 25403181 Free PMC article.
-
Identification of a noncanonical RNA binding domain in the U2 snRNP protein SF3A1.RNA. 2019 Nov;25(11):1509-1521. doi: 10.1261/rna.072256.119. Epub 2019 Aug 5. RNA. 2019. PMID: 31383795 Free PMC article.
-
Structural insights into recognition of SL4, the UUCG stem-loop, of human U1 snRNA by the ubiquitin-like domain, including the C-terminal tail in the SF3A1 subunit of U2 snRNP.J Biochem. 2023 Jul 31;174(2):203-216. doi: 10.1093/jb/mvad033. J Biochem. 2023. PMID: 37094335
-
RNA Splicing by the Spliceosome.Annu Rev Biochem. 2020 Jun 20;89:359-388. doi: 10.1146/annurev-biochem-091719-064225. Epub 2019 Dec 3. Annu Rev Biochem. 2020. PMID: 31794245 Review.
Cited by
-
Sequence-specific RNA recognition by an RGG motif connects U1 and U2 snRNP for spliceosome assembly.Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2114092119. doi: 10.1073/pnas.2114092119. Proc Natl Acad Sci U S A. 2022. PMID: 35101980 Free PMC article.
-
Inefficient recruitment of DDX39B impedes pre-spliceosome assembly on FOXP3 introns.RNA. 2024 Jun 17;30(7):824-838. doi: 10.1261/rna.079933.123. RNA. 2024. PMID: 38575347 Free PMC article.
-
Examining the capacity of human U1 snRNA variants to facilitate pre-mRNA splicing.RNA. 2024 Feb 16;30(3):271-280. doi: 10.1261/rna.079892.123. RNA. 2024. PMID: 38164604 Free PMC article.
-
Exon-independent recruitment of SRSF1 is mediated by U1 snRNP stem-loop 3.EMBO J. 2022 Jan 4;41(1):e107640. doi: 10.15252/embj.2021107640. Epub 2021 Nov 15. EMBO J. 2022. PMID: 34779515 Free PMC article.
-
Principles and correction of 5'-splice site selection.RNA Biol. 2022 Jan;19(1):943-960. doi: 10.1080/15476286.2022.2100971. RNA Biol. 2022. PMID: 35866748 Free PMC article. Review.
References
-
- Das R, Zhou Z, Reed R.. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell. 2000;5:779–787. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
