The role of RNA editing enzyme ADAR1 in human disease
- PMID: 34105255
- PMCID: PMC8651834
- DOI: 10.1002/wrna.1665
The role of RNA editing enzyme ADAR1 in human disease
Abstract
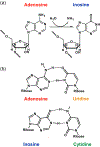
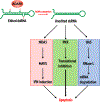
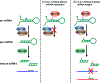
Adenosine deaminase acting on RNA (ADAR) catalyzes the posttranscriptional conversion of adenosine to inosine in double-stranded RNA (dsRNA), which can lead to the creation of missense mutations in coding sequences. Recent studies show that editing-dependent functions of ADAR1 protect dsRNA from dsRNA-sensing molecules and inhibit innate immunity and the interferon-mediated response. Deficiency in these ADAR1 functions underlie the pathogenesis of autoinflammatory diseases such as the type I interferonopathies Aicardi-Goutieres syndrome and dyschromatosis symmetrica hereditaria. ADAR1-mediated editing of endogenous coding and noncoding RNA as well as ADAR1 editing-independent interactions with DICER can also have oncogenic or tumor suppressive effects that affect tumor proliferation, invasion, and response to immunotherapy. The combination of proviral and antiviral roles played by ADAR1 in repressing the interferon response and editing viral RNAs alters viral morphogenesis and cell susceptibility to infection. This review analyzes the structure and function of ADAR1 with a focus on its position in human disease pathways and the mechanisms of its disease-associated effects. This article is categorized under: RNA in Disease and Development > RNA in Disease RNA Processing > RNA Editing and Modification RNA Interactions with Proteins and Other Molecules > Protein-RNA Interactions: Functional Implications.
Keywords: ADAR1; AGS; RNA editing; human disease; innate immunity.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article..
Figures

Similar articles
-
The RNA-editing enzyme ADAR1 controls innate immune responses to RNA.Cell Rep. 2014 Nov 20;9(4):1482-94. doi: 10.1016/j.celrep.2014.10.041. Epub 2014 Nov 13. Cell Rep. 2014. PMID: 25456137 Free PMC article.
-
Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown.Cell. 2018 Feb 8;172(4):811-824.e14. doi: 10.1016/j.cell.2017.12.038. Epub 2018 Jan 25. Cell. 2018. PMID: 29395325 Free PMC article.
-
Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutières-syndrome-like encephalopathy.Immunity. 2021 Sep 14;54(9):1976-1988.e7. doi: 10.1016/j.immuni.2021.08.022. Immunity. 2021. PMID: 34525338
-
ADAR RNA Modifications, the Epitranscriptome and Innate Immunity.Trends Biochem Sci. 2021 Sep;46(9):758-771. doi: 10.1016/j.tibs.2021.02.002. Epub 2021 Mar 15. Trends Biochem Sci. 2021. PMID: 33736931 Review.
-
RNA editing and immune control: from mechanism to therapy.Curr Opin Genet Dev. 2024 Jun;86:102195. doi: 10.1016/j.gde.2024.102195. Epub 2024 Apr 20. Curr Opin Genet Dev. 2024. PMID: 38643591 Review.
Cited by
-
ADAR2 enzymes: efficient site-specific RNA editors with gene therapy aspirations.RNA. 2022 Oct;28(10):1281-1297. doi: 10.1261/rna.079266.122. Epub 2022 Jul 21. RNA. 2022. PMID: 35863867 Free PMC article. Review.
-
Adenosine deaminase acting on RNA 1 (ADAR1) as crucial regulators in cardiovascular diseases: structures, pathogenesis, and potential therapeutic approach.Front Pharmacol. 2023 Aug 17;14:1194884. doi: 10.3389/fphar.2023.1194884. eCollection 2023. Front Pharmacol. 2023. PMID: 37663249 Free PMC article. Review.
-
Impact of Disease-Associated Mutations on the Deaminase Activity of ADAR1.Biochemistry. 2024 Feb 6;63(3):282-293. doi: 10.1021/acs.biochem.3c00405. Epub 2024 Jan 8. Biochemistry. 2024. PMID: 38190734 Free PMC article.
-
A pipeline for identifying guide RNA sequences that promote RNA editing of nonsense mutations that cause inherited retinal diseases.Mol Ther Nucleic Acids. 2024 Jan 30;35(1):102130. doi: 10.1016/j.omtn.2024.102130. eCollection 2024 Mar 12. Mol Ther Nucleic Acids. 2024. PMID: 38375504 Free PMC article.
-
Structural impacts of two disease-linked ADAR1 mutants: a molecular dynamics study.J Comput Aided Mol Des. 2024 Jul 17;38(1):25. doi: 10.1007/s10822-024-00565-1. J Comput Aided Mol Des. 2024. PMID: 39014124
References
FURTHER READING
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

