A comparison of five epidemiological models for transmission of SARS-CoV-2 in India
- PMID: 34098885
- PMCID: PMC8181542
- DOI: 10.1186/s12879-021-06077-9
A comparison of five epidemiological models for transmission of SARS-CoV-2 in India
Abstract
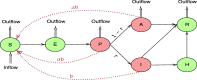
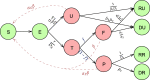
Background: Many popular disease transmission models have helped nations respond to the COVID-19 pandemic by informing decisions about pandemic planning, resource allocation, implementation of social distancing measures, lockdowns, and other non-pharmaceutical interventions. We study how five epidemiological models forecast and assess the course of the pandemic in India: a baseline curve-fitting model, an extended SIR (eSIR) model, two extended SEIR (SAPHIRE and SEIR-fansy) models, and a semi-mechanistic Bayesian hierarchical model (ICM).
Methods: Using COVID-19 case-recovery-death count data reported in India from March 15 to October 15 to train the models, we generate predictions from each of the five models from October 16 to December 31. To compare prediction accuracy with respect to reported cumulative and active case counts and reported cumulative death counts, we compute the symmetric mean absolute prediction error (SMAPE) for each of the five models. For reported cumulative cases and deaths, we compute Pearson's and Lin's correlation coefficients to investigate how well the projected and observed reported counts agree. We also present underreporting factors when available, and comment on uncertainty of projections from each model.
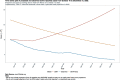
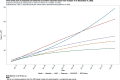
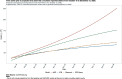
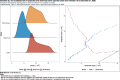
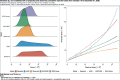
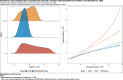
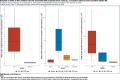
Results: For active case counts, SMAPE values are 35.14% (SEIR-fansy) and 37.96% (eSIR). For cumulative case counts, SMAPE values are 6.89% (baseline), 6.59% (eSIR), 2.25% (SAPHIRE) and 2.29% (SEIR-fansy). For cumulative death counts, the SMAPE values are 4.74% (SEIR-fansy), 8.94% (eSIR) and 0.77% (ICM). Three models (SAPHIRE, SEIR-fansy and ICM) return total (sum of reported and unreported) cumulative case counts as well. We compute underreporting factors as of October 31 and note that for cumulative cases, the SEIR-fansy model yields an underreporting factor of 7.25 and ICM model yields 4.54 for the same quantity. For total (sum of reported and unreported) cumulative deaths the SEIR-fansy model reports an underreporting factor of 2.97. On October 31, we observe 8.18 million cumulative reported cases, while the projections (in millions) from the baseline model are 8.71 (95% credible interval: 8.63-8.80), while eSIR yields 8.35 (7.19-9.60), SAPHIRE returns 8.17 (7.90-8.52) and SEIR-fansy projects 8.51 (8.18-8.85) million cases. Cumulative case projections from the eSIR model have the highest uncertainty in terms of width of 95% credible intervals, followed by those from SAPHIRE, the baseline model and finally SEIR-fansy.
Conclusions: In this comparative paper, we describe five different models used to study the transmission dynamics of the SARS-Cov-2 virus in India. While simulation studies are the only gold standard way to compare the accuracy of the models, here we were uniquely poised to compare the projected case-counts against observed data on a test period. The largest variability across models is observed in predicting the "total" number of infections including reported and unreported cases (on which we have no validation data). The degree of under-reporting has been a major concern in India and is characterized in this report. Overall, the SEIR-fansy model appeared to be a good choice with publicly available R-package and desired flexibility plus accuracy.
Keywords: Compartmental models; Low and middle income countries; Prediction uncertainty; Statistical models.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Extending the susceptible-exposed-infected-removed (SEIR) model to handle the false negative rate and symptom-based administration of COVID-19 diagnostic tests: SEIR-fansy.Stat Med. 2022 Jun 15;41(13):2317-2337. doi: 10.1002/sim.9357. Epub 2022 Feb 27. Stat Med. 2022. PMID: 35224743 Free PMC article.
-
Using outbreak data to estimate the dynamic COVID-19 landscape in Eastern Africa.BMC Infect Dis. 2022 Jun 9;22(1):531. doi: 10.1186/s12879-022-07510-3. BMC Infect Dis. 2022. PMID: 35681129 Free PMC article.
-
EXTENDING THE SUSCEPTIBLE-EXPOSED-INFECTED-REMOVED(SEIR) MODEL TO HANDLE THE HIGH FALSE NEGATIVE RATE AND SYMPTOM-BASED ADMINISTRATION OF COVID-19 DIAGNOSTIC TESTS: SEIR-fansy.medRxiv [Preprint]. 2020 Sep 25:2020.09.24.20200238. doi: 10.1101/2020.09.24.20200238. medRxiv. 2020. Update in: Stat Med. 2022 Jun 15;41(13):2317-2337. doi: 10.1002/sim.9357 PMID: 32995829 Free PMC article. Updated. Preprint.
-
Predictive model with analysis of the initial spread of COVID-19 in India.Int J Med Inform. 2020 Nov;143:104262. doi: 10.1016/j.ijmedinf.2020.104262. Epub 2020 Aug 25. Int J Med Inform. 2020. PMID: 32911257 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article. Review.
Cited by
-
Discrete Models in Epidemiology: New Contagion Probability Functions Based on Real Data Behavior.Bull Math Biol. 2022 Sep 22;84(11):127. doi: 10.1007/s11538-022-01076-6. Bull Math Biol. 2022. PMID: 36138179 Free PMC article.
-
Extending the susceptible-exposed-infected-removed (SEIR) model to handle the false negative rate and symptom-based administration of COVID-19 diagnostic tests: SEIR-fansy.Stat Med. 2022 Jun 15;41(13):2317-2337. doi: 10.1002/sim.9357. Epub 2022 Feb 27. Stat Med. 2022. PMID: 35224743 Free PMC article.
-
Monte Carlo simulation of COVID-19 pandemic using Planck's probability distribution.Biosystems. 2022 Aug;218:104708. doi: 10.1016/j.biosystems.2022.104708. Epub 2022 May 27. Biosystems. 2022. PMID: 35644321 Free PMC article.
-
Artificial intelligence for COVID-19: battling the pandemic with computational intelligence.Intell Med. 2022 Feb;2(1):13-29. doi: 10.1016/j.imed.2021.09.001. Epub 2021 Oct 21. Intell Med. 2022. PMID: 34697578 Free PMC article. Review.
-
Using outbreak data to estimate the dynamic COVID-19 landscape in Eastern Africa.BMC Infect Dis. 2022 Jun 9;22(1):531. doi: 10.1186/s12879-022-07510-3. BMC Infect Dis. 2022. PMID: 35681129 Free PMC article.
References
-
- Mayo Clinic . Coronavirus disease 2019 (COVID-19)—Symptoms and causes [Internet] 2020.
-
- Wikipedia . Coronavirus disease. 2019.
-
- Aiyar S. Covid-19 has exposed India’s failure to deliver even the most basic obligations to its people [Internet] CNN; 2020.
-
- Kulkarni S. India becomes third worst affected country by coronavirus, overtakes Russia Read more at: https://www.deccanherald.com/national/india-becomes-third-worst-affected... [Internet]. Deccan Herald. [cited 2020 Aug 3]. Available from: https://www.deccanherald.com/national/india-becomes-third-worst-affected....
-
- Basu D, Salvatore M, Ray D, Kleinsasser M, Purkayastha S, Bhattacharyya R, et al. A Comprehensive Public Health Evaluation of Lockdown as a Non-pharmaceutical Intervention on COVID-19 Spread in India: National Trends Masking State Level Variations [Internet]. Epidemiology. 2020; [cited 2020 Aug 3]. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.05.25.20113043. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

