Crumbs and the apical spectrin cytoskeleton regulate R8 cell fate in the Drosophila eye
- PMID: 34097697
- PMCID: PMC8211197
- DOI: 10.1371/journal.pgen.1009146
Crumbs and the apical spectrin cytoskeleton regulate R8 cell fate in the Drosophila eye
Abstract
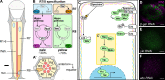
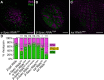
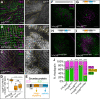
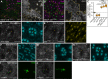
The Hippo pathway is an important regulator of organ growth and cell fate. In the R8 photoreceptor cells of the Drosophila melanogaster eye, the Hippo pathway controls the fate choice between one of two subtypes that express either the blue light-sensitive Rhodopsin 5 (Hippo inactive R8 subtype) or the green light-sensitive Rhodopsin 6 (Hippo active R8 subtype). The degree to which the mechanism of Hippo signal transduction and the proteins that mediate it are conserved in organ growth and R8 cell fate choice is currently unclear. Here, we identify Crumbs and the apical spectrin cytoskeleton as regulators of R8 cell fate. By contrast, other proteins that influence Hippo-dependent organ growth, such as the basolateral spectrin cytoskeleton and Ajuba, are dispensable for the R8 cell fate choice. Surprisingly, Crumbs promotes the Rhodopsin 5 cell fate, which is driven by Yorkie, rather than the Rhodopsin 6 cell fate, which is driven by Warts and the Hippo pathway, which contrasts with its impact on Hippo activity in organ growth. Furthermore, neither the apical spectrin cytoskeleton nor Crumbs appear to regulate the Hippo pathway through mechanisms that have been observed in growing organs. Together, these results show that only a subset of Hippo pathway proteins regulate the R8 binary cell fate decision and that aspects of Hippo signalling differ between growing organs and post-mitotic R8 cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The insulator protein BEAF-32 is required for Hippo pathway activity in the terminal differentiation of neuronal subtypes.Development. 2016 Jul 1;143(13):2389-97. doi: 10.1242/dev.134700. Epub 2016 May 25. Development. 2016. PMID: 27226322 Free PMC article.
-
The neuronal transcription factor erect wing regulates specification and maintenance of Drosophila R8 photoreceptor subtypes.Dev Biol. 2013 Sep 15;381(2):482-90. doi: 10.1016/j.ydbio.2013.07.001. Epub 2013 Jul 11. Dev Biol. 2013. PMID: 23850772 Free PMC article.
-
The Spectrin cytoskeleton regulates the Hippo signalling pathway.EMBO J. 2015 Apr 1;34(7):940-54. doi: 10.15252/embj.201489642. Epub 2015 Feb 23. EMBO J. 2015. PMID: 25712476 Free PMC article.
-
Upstream paths for Hippo signaling in Drosophila organ development.BMB Rep. 2018 Mar;51(3):134-142. doi: 10.5483/bmbrep.2018.51.3.027. BMB Rep. 2018. PMID: 29397870 Free PMC article. Review.
-
Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling.Dev Cell. 2012 Apr 17;22(4):695-702. doi: 10.1016/j.devcel.2012.03.013. Dev Cell. 2012. PMID: 22516196 Free PMC article. Review.
Cited by
-
The Hippo pathway uses different machinery to control cell fate and organ size.iScience. 2021 Jul 9;24(8):102830. doi: 10.1016/j.isci.2021.102830. eCollection 2021 Aug 20. iScience. 2021. PMID: 34355153 Free PMC article.
-
Protein Phosphatase 2A with B' specificity subunits regulates the Hippo-Yorkie signaling axis in the Drosophila eye disc.J Cell Sci. 2022 Oct 15;135(20):jcs259558. doi: 10.1242/jcs.259558. Epub 2022 Oct 25. J Cell Sci. 2022. PMID: 36205125 Free PMC article.
-
The ZO-1 protein Polychaetoid as an upstream regulator of the Hippo pathway in Drosophila.PLoS Genet. 2021 Nov 8;17(11):e1009894. doi: 10.1371/journal.pgen.1009894. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34748546 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

