RiboDoc: A Docker-based package for ribosome profiling analysis
- PMID: 34093996
- PMCID: PMC8141510
- DOI: 10.1016/j.csbj.2021.05.014
RiboDoc: A Docker-based package for ribosome profiling analysis
Abstract
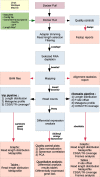
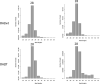
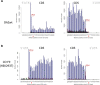
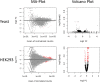
Ribosome profiling (RiboSeq) has emerged as a powerful technique for studying the genome-wide regulation of translation in various cells. Several steps in the biological protocol have been improved, but the bioinformatics part of RiboSeq suffers from a lack of standardization, preventing the straightforward and complete reproduction of published results. Too many published studies provide insufficient detail about the bioinformatics pipeline used. The broad range of questions that can be asked with RiboSeq makes it difficult to use a single bioinformatics tool. Indeed, many scripts have been published for addressing diverse questions. Here (https://github.com/equipeGST/RiboDoc), we propose a unique tool (for use with multiple operating systems, OS) to standardize the general steps that must be performed systematically in RiboSeq analysis, together with the statistical analysis and quality control of the sample. The data generated can then be exploited with more specific tools. We hope that this tool will help to standardize bioinformatics analyses pipelines in the field of translation.
Keywords: Bioinformatics; Docker; FAIR; OAZ1; Polyamines; Riboseq; Ribosome; Tool for ribosome analysis; Translation.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Riboseq-flow: A streamlined, reliable pipeline for ribosome profiling data analysis and quality control.Wellcome Open Res. 2024 Apr 11;9:179. doi: 10.12688/wellcomeopenres.21000.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 38846930 Free PMC article.
-
riboviz: analysis and visualization of ribosome profiling datasets.BMC Bioinformatics. 2017 Oct 25;18(1):461. doi: 10.1186/s12859-017-1873-8. BMC Bioinformatics. 2017. PMID: 29070028 Free PMC article.
-
Insights into the mechanisms of eukaryotic translation gained with ribosome profiling.Nucleic Acids Res. 2017 Jan 25;45(2):513-526. doi: 10.1093/nar/gkw1190. Epub 2016 Dec 6. Nucleic Acids Res. 2017. PMID: 27923997 Free PMC article. Review.
-
Shoelaces: an interactive tool for ribosome profiling processing and visualization.BMC Genomics. 2018 Jul 18;19(1):543. doi: 10.1186/s12864-018-4912-6. BMC Genomics. 2018. PMID: 30021517 Free PMC article.
-
Recent advances in ribosome profiling for deciphering translational regulation.Methods. 2020 Apr 1;176:46-54. doi: 10.1016/j.ymeth.2019.05.011. Epub 2019 May 17. Methods. 2020. PMID: 31103613 Review.
Cited by
-
A review of Ribosome profiling and tools used in Ribo-seq data analysis.Comput Struct Biotechnol J. 2024 Apr 22;23:1912-1918. doi: 10.1016/j.csbj.2024.04.051. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38721586 Free PMC article. Review.
-
2-Guanidino-quinazoline promotes the readthrough of nonsense mutations underlying human genetic diseases.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2122004119. doi: 10.1073/pnas.2122004119. Epub 2022 Aug 22. Proc Natl Acad Sci U S A. 2022. PMID: 35994666 Free PMC article.
-
Next generation sequencing technologies to address aberrant mRNA translation in cancer.NAR Cancer. 2024 May 15;6(2):zcae024. doi: 10.1093/narcan/zcae024. eCollection 2024 Jun. NAR Cancer. 2024. PMID: 38751936 Free PMC article. Review.
-
Riboseq-flow: A streamlined, reliable pipeline for ribosome profiling data analysis and quality control.Wellcome Open Res. 2024 Apr 11;9:179. doi: 10.12688/wellcomeopenres.21000.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 38846930 Free PMC article.
-
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.Plant Cell. 2024 Jul 31;36(8):2851-2872. doi: 10.1093/plcell/koae145. Plant Cell. 2024. PMID: 38723165
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
