The Roles of Host Noncoding RNAs in Mycobacterium tuberculosis Infection
- PMID: 34093557
- PMCID: PMC8170620
- DOI: 10.3389/fimmu.2021.664787
The Roles of Host Noncoding RNAs in Mycobacterium tuberculosis Infection
Abstract
Tuberculosis remains a major health problem. Mycobacterium tuberculosis, the causative agent of tuberculosis, can replicate and persist in host cells. Noncoding RNAs (ncRNAs) widely participate in various biological processes, including Mycobacterium tuberculosis infection, and play critical roles in gene regulation. In this review, we summarize the latest reports on ncRNAs (microRNAs, piRNAs, circRNAs and lncRNAs) that regulate the host response against Mycobacterium tuberculosis infection. In the context of host-Mycobacterium tuberculosis interactions, a broad and in-depth understanding of host ncRNA regulatory mechanisms may lead to potential clinical prospects for tuberculosis diagnosis and the development of new anti-tuberculosis therapies.
Keywords: Mycobacterium tuberculosis (M. tuberculosis); circRNA; immune response; lncRNA; miRNA; piRNA.
Copyright © 2021 Wei, Liu, Jia, Zhang, Bie and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
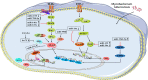
 Direct stimulatory modification;
Direct stimulatory modification;  Direct inhibitory modification.
Direct inhibitory modification.
 Direct stimulatory modification;
Direct stimulatory modification;  Direct inhibitory modification;
Direct inhibitory modification;  Tentative stimulatory modification;
Tentative stimulatory modification;  Tentative inhibitory modification.
Tentative inhibitory modification.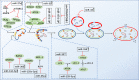
 Direct stimulatory modification;
Direct stimulatory modification;  Direct inhibitory modification;
Direct inhibitory modification;  Tentative stimulatory modification;
Tentative stimulatory modification;  Tentative inhibitory modification.
Tentative inhibitory modification.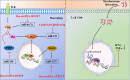
 Direct stimulatory modification;
Direct stimulatory modification;  Direct inhibitory modification;
Direct inhibitory modification;  Tentative stimulatory modification;
Tentative stimulatory modification;  Tentative inhibitory modification.
Tentative inhibitory modification.Similar articles
-
The Role of microRNAs and Long Non-Coding RNAs in the Regulation of the Immune Response to Mycobacterium tuberculosis Infection.Front Immunol. 2021 Jun 24;12:687962. doi: 10.3389/fimmu.2021.687962. eCollection 2021. Front Immunol. 2021. PMID: 34248974 Free PMC article. Review.
-
Long non-coding RNA molecules in tuberculosis.Int J Biol Macromol. 2020 Aug 1;156:340-346. doi: 10.1016/j.ijbiomac.2020.04.030. Epub 2020 Apr 10. Int J Biol Macromol. 2020. PMID: 32283111 Review.
-
Exosomal ncRNAs profiling of mycobacterial infection identified miRNA-185-5p as a novel biomarker for tuberculosis.Brief Bioinform. 2021 Nov 5;22(6):bbab210. doi: 10.1093/bib/bbab210. Brief Bioinform. 2021. PMID: 34169968
-
CD8 T cells and Mycobacterium tuberculosis infection.Semin Immunopathol. 2015 May;37(3):239-49. doi: 10.1007/s00281-015-0490-8. Epub 2015 Apr 28. Semin Immunopathol. 2015. PMID: 25917388 Free PMC article. Review.
-
MicroRNAs play big roles in modulating macrophages response toward mycobacteria infection.Infect Genet Evol. 2016 Nov;45:378-382. doi: 10.1016/j.meegid.2016.09.023. Epub 2016 Sep 28. Infect Genet Evol. 2016. PMID: 27693402 Review.
Cited by
-
Non-Coding RNAs in the Etiology and Control of Major and Neglected Human Tropical Diseases.Front Immunol. 2021 Oct 19;12:703936. doi: 10.3389/fimmu.2021.703936. eCollection 2021. Front Immunol. 2021. PMID: 34737736 Free PMC article. Review.
-
Non-Coding RNAs in Tuberculosis Epidemiology: Platforms and Approaches for Investigating the Genome's Dark Matter.Int J Mol Sci. 2022 Apr 17;23(8):4430. doi: 10.3390/ijms23084430. Int J Mol Sci. 2022. PMID: 35457250 Free PMC article. Review.
-
MicroRNA-155 Modulates Macrophages' Response to Non-Tuberculous Mycobacteria through COX-2/PGE2 Signaling.Pathogens. 2021 Jul 21;10(8):920. doi: 10.3390/pathogens10080920. Pathogens. 2021. PMID: 34451384 Free PMC article.
-
Long non-coding RNA expression in PBMCs of patients with active pulmonary tuberculosis.Front Microbiol. 2023 Dec 14;14:1257267. doi: 10.3389/fmicb.2023.1257267. eCollection 2023. Front Microbiol. 2023. PMID: 38156018 Free PMC article.
-
Long Non-Coding RNAs and Their "Discrete" Contribution to IBD and Johne's Disease-What Stands out in the Current Picture? A Comprehensive Review.Int J Mol Sci. 2023 Sep 1;24(17):13566. doi: 10.3390/ijms241713566. Int J Mol Sci. 2023. PMID: 37686376 Free PMC article. Review.
References
-
- World Health Organization. Global Tuberculosis Report. Geneva, Switzerland: World Health Organization; (2019).
-
- Wright A, Zignol M, Van Deun A, Falzon D, Gerdes SR, Feldman K, et al. . Epidemiology of Antituberculosis Drug Resistance 2002-07: An Updated Analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet (2009) 373(9678):1861–73. 10.1016/s0140-6736(09)60331-7 - DOI - PubMed
-
- MacDonald EM, Izzo AA. Tuberculosis Vaccine Development — Its History and Future Directions. In: Tuberculosis - Expanding Knowledge, IntechOpen (2015) 10.5772/59658 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

