Prognostic value of measurable residual disease monitoring by next-generation sequencing before and after allogeneic hematopoietic cell transplantation in acute myeloid leukemia
- PMID: 34088902
- PMCID: PMC8178334
- DOI: 10.1038/s41408-021-00500-9
Prognostic value of measurable residual disease monitoring by next-generation sequencing before and after allogeneic hematopoietic cell transplantation in acute myeloid leukemia
Abstract
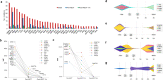
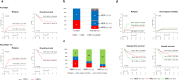
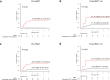
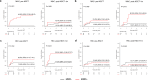
Given limited studies on next-generation sequencing-based measurable residual disease (NGS-MRD) in acute myeloid leukemia (AML) patients after allogeneic hematopoietic stem cell transplantation (allo-HSCT), we longitudinally collected samples before and after allo-HSCT from two independent prospective cohorts (n = 132) and investigated the prognostic impact of amplicon-based NGS assessment. Persistent mutations were detected pre-HSCT (43%) and 1 month after HSCT (post-HSCT-1m, 20%). All persistent mutations at both pre-HSCT and post-HSCT-1m were significantly associated with post-transplant relapse and worse overall survival. Changes in MRD status from pre-HSCT to post-HSCT-1m indicated a higher risk for relapse and death. Isolated detectable mutations in genes associated with clonal hematopoiesis were also significant predictors of post-transplant relapse. The optimal time point of NGS-MRD assessment depended on the conditioning intensity (pre-HSCT for myeloablative conditioning and post-HSCT-1m for reduced-intensity conditioning). Serial NGS-MRD monitoring revealed that most residual clones at both pre-HSCT and post-HSCT-1m in patients who never relapsed disappeared after allo-HSCT. Reappearance of mutant clones before overt relapse was detected by the NGS-MRD assay. Taken together, NGS-MRD detection has a prognostic value at both pre-HSCT and post-HSCT-1m, regardless of the mutation type, depending on the conditioning intensity. Serial NGS-MRD monitoring was feasible to compensate for the limited performance of the NGS-MRD assay.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Next-generation sequencing-defined minimal residual disease before stem cell transplantation predicts acute myeloid leukemia relapse.Am J Hematol. 2019 Aug;94(8):902-912. doi: 10.1002/ajh.25514. Epub 2019 Jun 14. Am J Hematol. 2019. PMID: 31124175
-
WT1 Measurable Residual Disease Assay in Patients With Acute Myeloid Leukemia Who Underwent Allogeneic Hematopoietic Stem Cell Transplantation: Optimal Time Points, Thresholds, and Candidates.Biol Blood Marrow Transplant. 2019 Oct;25(10):1925-1932. doi: 10.1016/j.bbmt.2019.05.033. Epub 2019 Jun 4. Biol Blood Marrow Transplant. 2019. PMID: 31173897
-
Impact of Conditioning Intensity of Allogeneic Transplantation for Acute Myeloid Leukemia With Genomic Evidence of Residual Disease.J Clin Oncol. 2020 Apr 20;38(12):1273-1283. doi: 10.1200/JCO.19.03011. Epub 2019 Dec 20. J Clin Oncol. 2020. PMID: 31860405 Free PMC article.
-
Next-generation sequencing-based minimal residual disease monitoring in patients receiving allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia or myelodysplastic syndrome.Curr Opin Hematol. 2018 Nov;25(6):425-432. doi: 10.1097/MOH.0000000000000464. Curr Opin Hematol. 2018. PMID: 30281033 Review.
-
The Role of Measurable Residual Disease (MRD) in Hematopoietic Stem Cell Transplantation for Hematological Malignancies Focusing on Acute Leukemia.Int J Mol Sci. 2019 Oct 28;20(21):5362. doi: 10.3390/ijms20215362. Int J Mol Sci. 2019. PMID: 31661875 Free PMC article. Review.
Cited by
-
New drugs before, during, and after hematopoietic stem cell transplantation for patients with acute myeloid leukemia.Haematologica. 2023 Feb 1;108(2):321-341. doi: 10.3324/haematol.2022.280798. Haematologica. 2023. PMID: 36722403 Free PMC article. Review.
-
Depth of Response to Intensive Chemotherapy Has Significant Prognostic Value among Acute Myeloid Leukemia (AML) Patients Undergoing Allogeneic Hematopoietic Stem-Cell Transplantation with Intermediate or Adverse Risk at Diagnosis Compared to At-Risk Group According to European Leukemia Net 2017 Risk Stratification.Cancers (Basel). 2022 Jun 29;14(13):3199. doi: 10.3390/cancers14133199. Cancers (Basel). 2022. PMID: 35804971 Free PMC article.
-
Molecular Minimal Residual Disease Detection in Acute Myeloid Leukemia.Cancers (Basel). 2021 Oct 29;13(21):5431. doi: 10.3390/cancers13215431. Cancers (Basel). 2021. PMID: 34771594 Free PMC article. Review.
-
FLT3-ITD Measurable Residual Disease Monitoring in Acute Myeloid Leukemia Using Next-Generation Sequencing.Cancers (Basel). 2022 Dec 12;14(24):6121. doi: 10.3390/cancers14246121. Cancers (Basel). 2022. PMID: 36551616 Free PMC article.
-
Dynamic assessment of measurable residual disease in favorable-risk acute myeloid leukemia in first remission, treatment, and outcomes.Blood Cancer J. 2021 Dec 6;11(12):195. doi: 10.1038/s41408-021-00591-4. Blood Cancer J. 2021. PMID: 34873148 Free PMC article.
References
-
- Bejanyan N, et al. Survival of patients with acute myeloid leukemia relapsing after allogeneic hematopoietic cell transplantation: a center for international blood and marrow transplant research study. Biol. Blood Marrow Transplant. 2015;21:454–459. doi: 10.1016/j.bbmt.2014.11.007. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

