Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens
- PMID: 34079067
- PMCID: PMC8636523
- DOI: 10.1038/s41380-021-01163-5
Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens
Abstract
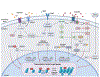
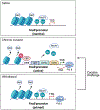
Repeated cocaine use induces coordinated changes in gene expression that drive plasticity in the nucleus accumbens (NAc), an important component of the brain's reward circuitry, and promote the development of maladaptive, addiction-like behaviors. Studies on the molecular basis of cocaine action identify transcription factors, a class of proteins that bind to specific DNA sequences and regulate transcription, as critical mediators of this cocaine-induced plasticity. Early methods to identify and study transcription factors involved in addiction pathophysiology primarily relied on quantifying the expression of candidate genes in bulk brain tissue after chronic cocaine treatment, as well as conventional overexpression and knockdown techniques. More recently, advances in next generation sequencing, bioinformatics, cell-type-specific targeting, and locus-specific neuroepigenomic editing offer a more powerful, unbiased toolbox to identify the most important transcription factors that drive drug-induced plasticity and to causally define their downstream molecular mechanisms. Here, we synthesize the literature on transcription factors mediating cocaine action in the NAc, discuss the advancements and remaining limitations of current experimental approaches, and emphasize recent work leveraging bioinformatic tools and neuroepigenomic editing to study transcription factors involved in cocaine addiction.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Conflicts of interest:
None.
Figures

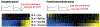
Similar articles
-
HDAC3 Activity within the Nucleus Accumbens Regulates Cocaine-Induced Plasticity and Behavior in a Cell-Type-Specific Manner.J Neurosci. 2021 Mar 31;41(13):2814-2827. doi: 10.1523/JNEUROSCI.2829-20.2021. Epub 2021 Feb 18. J Neurosci. 2021. PMID: 33602824 Free PMC article.
-
Circuit-Wide Gene Network Analysis Reveals Sex-Specific Roles for Phosphodiesterase 1b in Cocaine Addiction.J Neurosci. 2024 Jun 5;44(23):e1327232024. doi: 10.1523/JNEUROSCI.1327-23.2024. J Neurosci. 2024. PMID: 38637154
-
Cocaine-induced metaplasticity in the nucleus accumbens: silent synapse and beyond.Neuropharmacology. 2011 Dec;61(7):1060-9. doi: 10.1016/j.neuropharm.2010.12.033. Epub 2011 Jan 11. Neuropharmacology. 2011. PMID: 21232547 Free PMC article. Review.
-
Transcription Factor E2F3a in Nucleus Accumbens Affects Cocaine Action via Transcription and Alternative Splicing.Biol Psychiatry. 2018 Aug 1;84(3):167-179. doi: 10.1016/j.biopsych.2017.11.027. Epub 2017 Dec 5. Biol Psychiatry. 2018. PMID: 29397901 Free PMC article.
-
Regulation of AMPA receptor trafficking in the nucleus accumbens by dopamine and cocaine.Neurotox Res. 2010 Nov;18(3-4):393-409. doi: 10.1007/s12640-010-9176-0. Epub 2010 Apr 2. Neurotox Res. 2010. PMID: 20361291 Free PMC article. Review.
Cited by
-
Multi-omics profiling of DNA methylation and gene expression alterations in human cocaine use disorder.Transl Psychiatry. 2024 Oct 9;14(1):428. doi: 10.1038/s41398-024-03139-9. Transl Psychiatry. 2024. PMID: 39384764 Free PMC article.
-
Cell type-specific epigenetic priming of gene expression in nucleus accumbens by cocaine.Sci Adv. 2024 Oct 4;10(40):eado3514. doi: 10.1126/sciadv.ado3514. Epub 2024 Oct 4. Sci Adv. 2024. PMID: 39365860 Free PMC article.
-
GAN-WGCNA: Calculating gene modules to identify key intermediate regulators in cocaine addiction.PLoS One. 2024 Oct 3;19(10):e0311164. doi: 10.1371/journal.pone.0311164. eCollection 2024. PLoS One. 2024. PMID: 39361596 Free PMC article.
-
Elucidating the molecular symphony: unweaving the transcriptional & epigenetic pathways underlying neuroplasticity in opioid dependence and withdrawal.Psychopharmacology (Berl). 2024 Oct;241(10):1955-1981. doi: 10.1007/s00213-024-06684-9. Epub 2024 Sep 10. Psychopharmacology (Berl). 2024. PMID: 39254835 Review.
-
The nucleus accumbens in reward and aversion processing: insights and implications.Front Behav Neurosci. 2024 Aug 9;18:1420028. doi: 10.3389/fnbeh.2024.1420028. eCollection 2024. Front Behav Neurosci. 2024. PMID: 39184934 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

