Setd4 controlled quiescent c-Kit+ cells contribute to cardiac neovascularization of capillaries beyond activation
- PMID: 34079011
- PMCID: PMC8172824
- DOI: 10.1038/s41598-021-91105-6
Setd4 controlled quiescent c-Kit+ cells contribute to cardiac neovascularization of capillaries beyond activation
Abstract
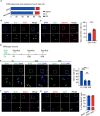
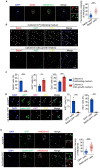
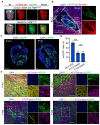
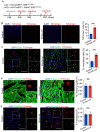
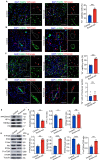
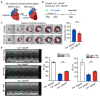
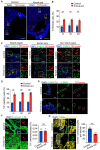
Blood vessels in the adult mammal exist in a highly organized and stable state. In the ischemic heart, limited expansion capacity of the myocardial vascular bed cannot satisfy demands for oxygen supply and the myocardium eventually undergoes irreversible damage. The predominant contribution of endogenous c-Kit+ cells is understood to be in the development and homeostasis of cardiac endothelial cells, which suggests potential for their targeting in treatments for cardiac ischemic injury. Quiescent cells in other tissues are known to contribute to the long-term maintenance of a cell pool, preserve proliferation capacity and, upon activation, facilitate tissue homeostasis and regeneration in response to tissue injury. Here, we present evidence of a Setd4-expressing quiescent c-Kit+ cell population in the adult mouse heart originating from embryonic stages. Conditional knock-out of Setd4 in c-Kit-CreERT2;Setd4f/f;Rosa26TdTomato mice induced an increase in vascular endothelial cells of capillaries in both neonatal and adult mice. We show that Setd4 regulates quiescence of c-Kit+ cells by the PI3K-Akt-mTOR signaling pathway via H4K20me3 catalysis. In myocardial infarction injured mice, Setd4 knock-out resulted in attenuated cardiomyocyte apoptosis, decreased infarction size and improved cardiac function. Lineage tracing in Setd4-Cre;Rosa26mT/mG mice showed that Setd4+ cells contribute to each cardiac lineage. Overall, Setd4 epigenetically controls c-Kit+ cell quiescence in the adult heart by facilitating heterochromatin formation via H4K20me3. Beyond activation, endogenous quiescent c-Kit+ cells were able to improve cardiac function in myocardial infarction injured mice via the neovascularization of capillaries.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
SET Domain-Containing Protein 4 Epigenetically Controls Breast Cancer Stem Cell Quiescence.Cancer Res. 2019 Sep 15;79(18):4729-4743. doi: 10.1158/0008-5472.CAN-19-1084. Epub 2019 Jul 15. Cancer Res. 2019. PMID: 31308046
-
Gata4-Dependent Differentiation of c-Kit+-Derived Endothelial Cells Underlies Artefactual Cardiomyocyte Regeneration in the Heart.Circulation. 2018 Sep 4;138(10):1012-1024. doi: 10.1161/CIRCULATIONAHA.118.033703. Circulation. 2018. PMID: 29666070 Free PMC article.
-
Nerve growth factor promotes cardiac repair following myocardial infarction.Circ Res. 2010 Apr 16;106(7):1275-84. doi: 10.1161/CIRCRESAHA.109.210088. Epub 2010 Apr 1. Circ Res. 2010. PMID: 20360245 Free PMC article.
-
An overview of the myocardial regeneration potential of cardiac c-Kit+ progenitor cells via PI3K and MAPK signaling pathways.Future Cardiol. 2020 May;16(3):199-209. doi: 10.2217/fca-2018-0049. Epub 2020 Mar 3. Future Cardiol. 2020. PMID: 32125173 Review.
-
c-kit(+) cells: the tell-tale heart of cardiac regeneration?Cell Mol Life Sci. 2015 May;72(9):1725-40. doi: 10.1007/s00018-014-1832-8. Epub 2015 Jan 10. Cell Mol Life Sci. 2015. PMID: 25575564 Free PMC article. Review.
Cited by
-
Recent Insights into Endogenous Mammalian Cardiac Regeneration Post-Myocardial Infarction.Int J Mol Sci. 2024 Nov 1;25(21):11747. doi: 10.3390/ijms252111747. Int J Mol Sci. 2024. PMID: 39519298 Free PMC article. Review.
-
Modified Taohong Siwu decoction improves cardiac function after myocardial ischaemia and reperfusion in rats by promoting endogenous stem cell mobilization and regulating metabolites.Pharm Biol. 2022 Dec;60(1):1721-1731. doi: 10.1080/13880209.2022.2116054. Pharm Biol. 2022. PMID: 36086864 Free PMC article.
-
Interactions between the ERK1/2 signaling pathway and PCAF play a key role in PE‑induced cardiomyocyte hypertrophy.Mol Med Rep. 2021 Sep;24(3):636. doi: 10.3892/mmr.2021.12275. Epub 2021 Jul 19. Mol Med Rep. 2021. PMID: 34278478 Free PMC article.
-
Unmasking the mammalian SET domain-containing protein 4.NAR Cancer. 2022 Jul 13;4(3):zcac021. doi: 10.1093/narcan/zcac021. eCollection 2022 Sep. NAR Cancer. 2022. PMID: 35854936 Free PMC article.
-
The Vascular Niche for Adult Cardiac Progenitor Cells.Antioxidants (Basel). 2022 Apr 29;11(5):882. doi: 10.3390/antiox11050882. Antioxidants (Basel). 2022. PMID: 35624750 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

