Discovery and Characterization of Actively Replicating DNA and Retro-Transcribing Viruses in Lower Vertebrate Hosts Based on RNA Sequencing
- PMID: 34072878
- PMCID: PMC8227577
- DOI: 10.3390/v13061042
Discovery and Characterization of Actively Replicating DNA and Retro-Transcribing Viruses in Lower Vertebrate Hosts Based on RNA Sequencing
Abstract
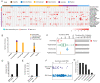
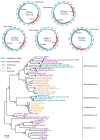
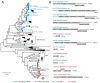
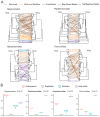
In a previous study, a metatranscriptomics survey of RNA viruses in several important lower vertebrate host groups revealed huge viral diversity, transforming the understanding of the evolution of vertebrate-associated RNA virus groups. However, the diversity of the DNA and retro-transcribing viruses in these host groups was left uncharacterized. Given that RNA sequencing is capable of revealing viruses undergoing active transcription and replication, we collected previously generated datasets associated with lower vertebrate hosts, and searched them for DNA and retro-transcribing viruses. Our results revealed the complete genome, or "core gene sets", of 18 vertebrate-associated DNA and retro-transcribing viruses in cartilaginous fishes, ray-finned fishes, and amphibians, many of which had high abundance levels, and some of which showed systemic infections in multiple organs, suggesting active transcription or acute infection within the host. Furthermore, these new findings recharacterized the evolutionary history in the families Hepadnaviridae, Papillomaviridae, and Alloherpesviridae, confirming long-term virus-host codivergence relationships for these virus groups. Collectively, our results revealed reliable and sufficient information within metatranscriptomics sequencing to characterize not only RNA viruses, but also DNA and retro-transcribing viruses, and therefore established a key methodology that will help us to understand the composition and evolution of the total "infectome" within a diverse range of vertebrate hosts.
Keywords: codivergence; metatranscriptomics; vertebrate-associated DNA virus; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The evolutionary history of vertebrate RNA viruses.Nature. 2018 Apr;556(7700):197-202. doi: 10.1038/s41586-018-0012-7. Epub 2018 Apr 4. Nature. 2018. PMID: 29618816
-
Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer.Virus Res. 2018 Jan 15;244:36-52. doi: 10.1016/j.virusres.2017.10.020. Epub 2017 Nov 8. Virus Res. 2018. PMID: 29103997 Free PMC article. Review.
-
Phytovirome Analysis of Wild Plant Populations: Comparison of Double-Stranded RNA and Virion-Associated Nucleic Acid Metagenomic Approaches.J Virol. 2019 Dec 12;94(1):e01462-19. doi: 10.1128/JVI.01462-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597769 Free PMC article.
-
High-Resolution Metatranscriptomics Reveals the Ecological Dynamics of Mosquito-Associated RNA Viruses in Western Australia.J Virol. 2017 Aug 10;91(17):e00680-17. doi: 10.1128/JVI.00680-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637756 Free PMC article.
-
The diversity, evolution and origins of vertebrate RNA viruses.Curr Opin Virol. 2018 Aug;31:9-16. doi: 10.1016/j.coviro.2018.07.017. Epub 2018 Aug 13. Curr Opin Virol. 2018. PMID: 30114593 Free PMC article. Review.
Cited by
-
First Isolation of a Herpesvirus (Family Alloherpesviridae) from Great Lakes Lake Sturgeon (Acipenser fulvescens).Animals (Basel). 2022 Nov 22;12(23):3230. doi: 10.3390/ani12233230. Animals (Basel). 2022. PMID: 36496751 Free PMC article.
-
Revealing the uncharacterised diversity of amphibian and reptile viruses.ISME Commun. 2022 Oct 2;2(1):95. doi: 10.1038/s43705-022-00180-x. ISME Commun. 2022. PMID: 37938670 Free PMC article.
-
Virome Analysis of Normal and Growth Retardation Disease-Affected Macrobrachium rosenbergii.Microbiol Spectr. 2022 Dec 21;10(6):e0146222. doi: 10.1128/spectrum.01462-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445118 Free PMC article.
-
Structural conservation of HBV-like capsid proteins over hundreds of millions of years despite the shift from non-enveloped to enveloped life-style.Nat Commun. 2023 Mar 22;14(1):1574. doi: 10.1038/s41467-023-37068-w. Nat Commun. 2023. PMID: 36949039 Free PMC article.
-
A parasite odyssey: An RNA virus concealed in Toxoplasma gondii.Virus Evol. 2024 May 11;10(1):veae040. doi: 10.1093/ve/veae040. eCollection 2024. Virus Evol. 2024. PMID: 38817668 Free PMC article.
References
-
- Cook S., Chung B.Y., Bass D., Moureau G., Tang S., McAlister E., Culverwell C.L., Glücksman E., Wang H., Brown T.D., et al. Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts. PLoS ONE. 2013;8:e80720. doi: 10.1371/journal.pone.0080720. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

