H3K4me2 Promotes the Activation of lncCPSET1 by Jun in the Chicken PGC Formation
- PMID: 34072197
- PMCID: PMC8227976
- DOI: 10.3390/ani11061572
H3K4me2 Promotes the Activation of lncCPSET1 by Jun in the Chicken PGC Formation
Abstract
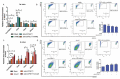
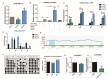
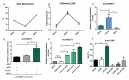
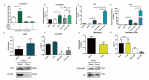
Primordial germ cells are the ancestors of female and male cells. Current research has shown that long non-coding RNA (lncRNA) and Histone methylation are the pivotal epigenetic factors in the PGC formation. However, there are few studies on the regulatory mechanism of lncRNA in the formation of PGC. Here, we define the lncRNA highly expressed in chicken PGC, lncCPSET1 (chicken-PGC-specifically-expressed transcript 1) This study found that compared with the interference of lncCPSET1/histone methylase Mll2 alone, the PGC formation was severely inhibited with the interference of lncCPSET1 and histone methylase Mll2 jointly in vivo and in vitro. Studies on the transcription level of lncCPSET1 found that H3K4me2 and transcription factor Jun have a positive effect on the activation of lncCPSET1; while DNA hypomethylation inhibits the expression of lncCPSET1. In terms of mechanism, compared with DNA methylation, H3K4me2 dominates lncCPSET1 activation. H3K4me2 can be enriched in the lncCPSET1 promoter, change its chromosome conformation, recruit the transcription factor Jun, and activate the expression of lncCPSET1. Taken together, we confirmed the model that H3K4me2 rather than DNA hypomethylation mediates Jun to regulate lncCPSET1 transcription, which broadens the study of lncCPSET1 pre-transcriptional mechanism.
Keywords: DNA methylation; H3K4me2; chicken; lncRNA; primordial germ cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
lncCPSET1 acts as a scaffold for MLL2/COMPASS to regulate Bmp4 and promote the formation of chicken primordial germ cells.Mol Genet Genomics. 2024 Mar 29;299(1):41. doi: 10.1007/s00438-024-02127-4. Mol Genet Genomics. 2024. PMID: 38551742
-
Narrow H3K4me2 is required for chicken PGC formation.J Cell Physiol. 2021 Feb;236(2):1391-1400. doi: 10.1002/jcp.29945. Epub 2020 Aug 4. J Cell Physiol. 2021. PMID: 32749682
-
P53 and H3K4me2 activate N6-methylated LncPGCAT-1 to regulate primordial germ cell formation via MAPK signaling.J Cell Physiol. 2020 Dec;235(12):9895-9909. doi: 10.1002/jcp.29805. Epub 2020 May 27. J Cell Physiol. 2020. PMID: 32458486
-
A current view of the epigenome in mouse primordial germ cells.Mol Reprod Dev. 2014 Feb;81(2):160-70. doi: 10.1002/mrd.22214. Epub 2013 Aug 7. Mol Reprod Dev. 2014. PMID: 23868517 Review.
-
Establishment and inheritance of epigenetic transcriptional memory.Front Mol Biosci. 2022 Sep 2;9:977653. doi: 10.3389/fmolb.2022.977653. eCollection 2022. Front Mol Biosci. 2022. PMID: 36120540 Free PMC article. Review.
Cited by
-
Ten-eleven translocation 1 mediating DNA demethylation regulates the proliferation of chicken primordial germ cells through the activation of Wnt4/β-catenin signaling pathway.Anim Biosci. 2024 Mar;37(3):471-480. doi: 10.5713/ab.23.0310. Epub 2024 Jan 20. Anim Biosci. 2024. PMID: 38271970 Free PMC article.
-
Inhibition of Autophagy Maintains ESC Pluripotency and Inhibits Primordial Germ Cell Formation in Chickens.Stem Cells Int. 2023 Apr 4;2023:4956871. doi: 10.1155/2023/4956871. eCollection 2023. Stem Cells Int. 2023. PMID: 37056458 Free PMC article.
-
Fate Decisions of Chicken Primordial Germ Cells (PGCs): Development, Integrity, Sex Determination, and Self-Renewal Mechanisms.Genes (Basel). 2023 Feb 28;14(3):612. doi: 10.3390/genes14030612. Genes (Basel). 2023. PMID: 36980885 Free PMC article. Review.
References
-
- Wang M., Zhang C., Huang C., Cheng S., He N., Wang Y., Ahmed M.F., Zhao R., Jin J., Zuo Q., et al. Regulation of fibroblast growth factor 8 (FGF8) in chicken embryonic stem cells differentiation into spermatogonial stem cells. J. Cell. Biochem. 2018;119:2396–2407. doi: 10.1002/jcb.26402. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

